
Links to IEMbase and Treatable ID have moved – they can now be found on the new Therapies tab
28.01.2026 12:49 — 👍 2 🔁 1 💬 0 📌 0@deciphergenomics.bsky.social

Links to IEMbase and Treatable ID have moved – they can now be found on the new Therapies tab
28.01.2026 12:49 — 👍 2 🔁 1 💬 0 📌 0
Approved genetic drugs/therapies, from the @n1collaborative.bsky.social, N1C Gene Registry are now displayed in DECIPHER on a new Therapies tab - available from gene pages and patient records.
28.01.2026 12:48 — 👍 2 🔁 1 💬 0 📌 0
Links to @uniquecharity.bsky.social single gene disorder guides are now displayed in DECIPHER on gene pages, therapies tabs and patient records #inclusion #informationforeveryone
28.01.2026 12:47 — 👍 2 🔁 1 💬 0 📌 1DECIPHER version 11.37 has been released. See the new features at www.deciphergenomics.org #variantinterpretation
28.01.2026 12:46 — 👍 1 🔁 1 💬 0 📌 0
I'm excited to be speaking at #FOGLondon this January.
Have a question you'd like me to answer during my session? Drop it in the comments or message me, I'd love to hear your thoughts.
Further information: hubs.la/Q03JMvwd0
#FOGLondon #genomics #biodata

Compiling evidence of gene-disease associations from the scientific literature for rare disease diagnosis and research is essential but time consuming.
Publications identified using a new machine learning approach can now be searched and browsed in G2P.
www.ebi.ac.uk/about/news/u...

On the protein browser mtDNA @gnomad-project.bsky.social missense and LoF tracks are available which display the location of gnomAD variants with these predicted molecular consequences.
31.10.2025 10:09 — 👍 3 🔁 0 💬 0 📌 0
On the genome browser, @gnomad-project.bsky.social mitochondrial variants and coverage tracks are now available. Variants can be coloured by predicted consequence, homoplasmic allele frequency or heteroplasmic allele frequency.
31.10.2025 10:08 — 👍 1 🔁 0 💬 0 📌 0
On annotation tabs, @gnomad-project.bsky.social mitochondrial DNA variant homoplasmic and heteroplasmic counts and allele frequencies for each haplogroup are displayed alongside lineage information from MITOMAP. Heteroplasmy distribution and coverage metrics are also available
31.10.2025 10:07 — 👍 3 🔁 0 💬 0 📌 0DECIPHER version 11.35 has been released. See the new features at www.deciphergenomics.org #variantinterpretation
31.10.2025 10:07 — 👍 2 🔁 1 💬 0 📌 0
Growth charts guide child healthcare, but standard charts often don’t reflect the growth patterns of children with rare conditions.
A new method, LMSz, creates condition-specific growth charts and is being integrated in @deciphergenomics.bsky.social
www.ebi.ac.uk/about/news/t...
🧬💻

Super-simple application of pathogenicity evidence during variant assessment in @deciphergenomics.bsky.social - even for complicated PVS1 in multiexon deletions where the frame is preserved - confirming a likely diagnosis.
11.09.2025 08:25 — 👍 7 🔁 2 💬 0 📌 0
Very cool new feature in @deciphergenomics.bsky.social - direct link from any missense variant to ProtVar @ebi.embl.org. This variant is in the binding site and likely interacts with the ligand, predicted using AlphaFold with AlphaFill!
10.09.2025 09:54 — 👍 13 🔁 4 💬 0 📌 0
The bespoke PubMed search on gene pages is now displayed in bold. This link opens a browser tab with a PubMed search displaying publications that include the gene of interest.
10.09.2025 15:50 — 👍 3 🔁 0 💬 0 📌 1
Descriptive names for gene and protein predictive scores are now displayed on gene pages to assist in demystifying these scores and making them easier to understand.
10.09.2025 15:50 — 👍 1 🔁 0 💬 0 📌 0
ClinGen Variant Curation Expert Panel Recommendations are now displayed more clearly on gene pages and in the pathogenicity evidence interface, especially for genes with recommendations for more than one disease @theacmg.bsky.social
10.09.2025 15:49 — 👍 2 🔁 1 💬 0 📌 0
Links to ProtVar are now available from the protein browser which provide functional and structural annotations for missense variants @ebi.embl.org
10.09.2025 15:49 — 👍 3 🔁 0 💬 0 📌 1
Mitochondrial gene predictive scores are now displayed on gene pages which indicate the observed depletion or enrichment of specific variants classes in @gnomad-project.bsky.social l compared to a mitochondrial genome constraint model under neutrality selection.
10.09.2025 15:48 — 👍 1 🔁 0 💬 0 📌 0DECIPHER version 11.34 has been released. See the new features at www.deciphergenomics.org #variantinterpretation
10.09.2025 15:47 — 👍 4 🔁 1 💬 0 📌 2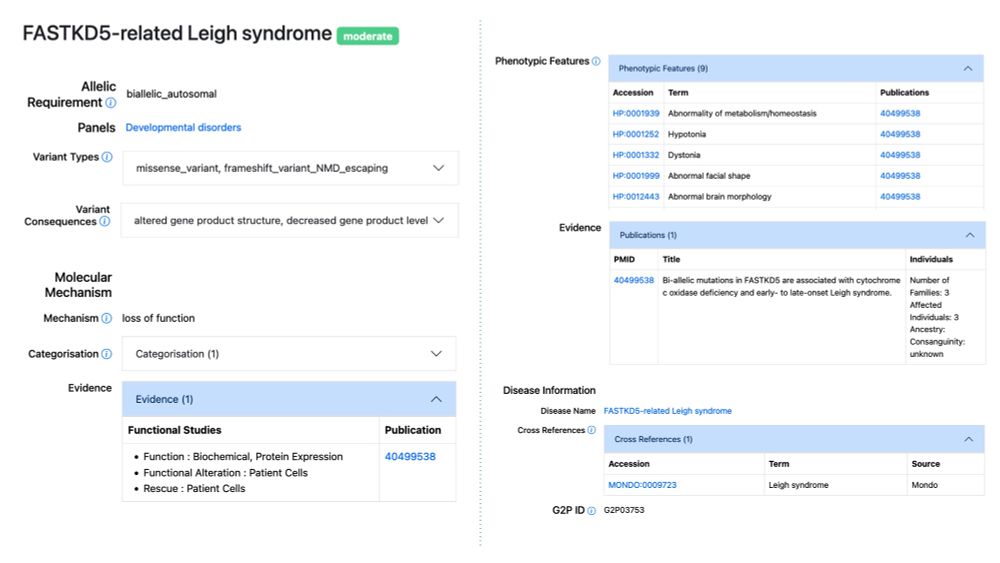
FASTKD5-related Leigh syndrome added to DDG2P. Biallelic FASTKD5 LoF variants cause an early- to late-onset Leigh syndrome associated with complex IV deficiency. www.ebi.ac.uk/gene2phenoty...
17.07.2025 09:54 — 👍 2 🔁 1 💬 0 📌 0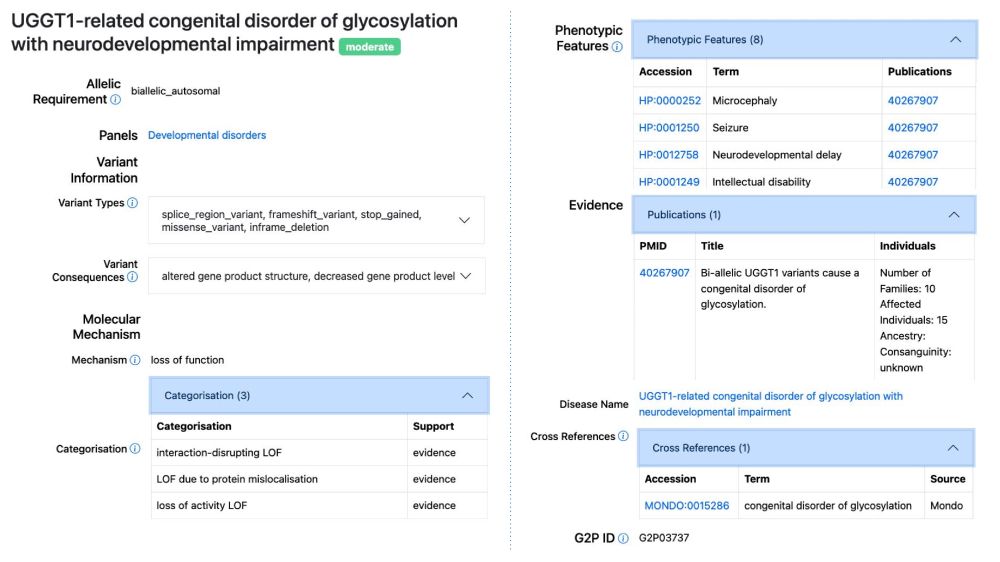
'UGGT1-related congenital disorder of glycosylation with neurodevelopmental impairment' added to DDG2P. Biallelic UGGT1 LoF variants cause a disorder characterised by developmental delay, intellectual disability, seizures, craniofacial dysmorphism, and microcephaly. See www.ebi.ac.uk/gene2phenoty...
11.07.2025 09:19 — 👍 2 🔁 1 💬 0 📌 0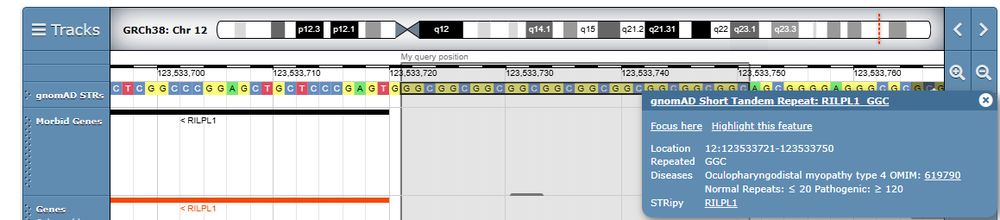
18 additional @gnomad-project.bsky.social short tandem repeats are now displayed; 9 recently discovered and 9 which are not currently linked to rare diseases but have historically appeared in various catalogs as suspected disease-causing loci.
10.07.2025 09:07 — 👍 0 🔁 0 💬 0 📌 0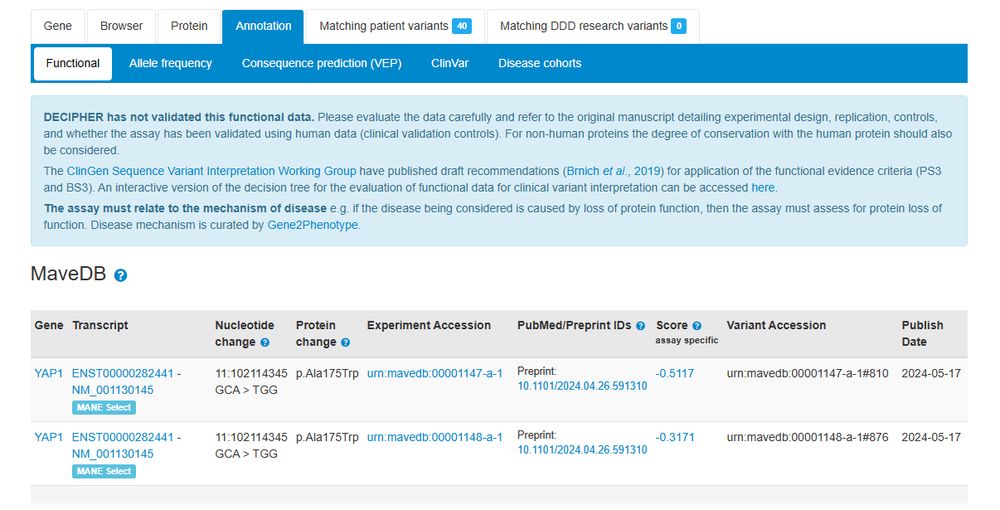
Additional functional data from Multiplexed Assays of Variant Effect (MAVEs) are now displayed on functional tabs. Previously only published datasets were displayed, now datasets with a preprint are available @varianteffect.bsky.social
10.07.2025 09:06 — 👍 2 🔁 0 💬 0 📌 0DECIPHER version 11.33 has been released. See the new features at www.deciphergenomics.org #variantinterpretation
10.07.2025 09:05 — 👍 1 🔁 0 💬 0 📌 0
Wellcome Connecting Science Interpreting Genomic Variation: Overcoming Challenges in Diverse Populations Free online course Starts on 7 July Live for three weeks Sign up today
Unravel the complexities of genome variation with our FREE online course! #FLGenomicVariantsDiversity
🗓️ Starts: 7 July
Learn how to get the most from #genomics data from diverse populations, using variant classification and interpretation approaches. #GenomeVariation🧬
📎bit.ly/3R7G7aK
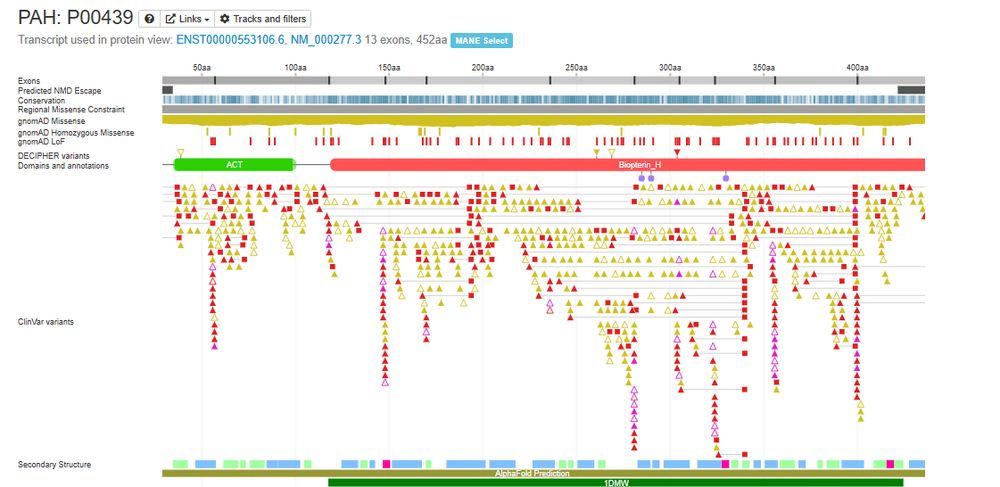
DECIPHER and #clinvar variants with a predicted molecular consequence of splice_donor_region_variant and splice_polypyrimidine_tract_variant are now displayed on the protein browser. These are displayed as pink triangles.
18.06.2025 14:26 — 👍 4 🔁 2 💬 0 📌 0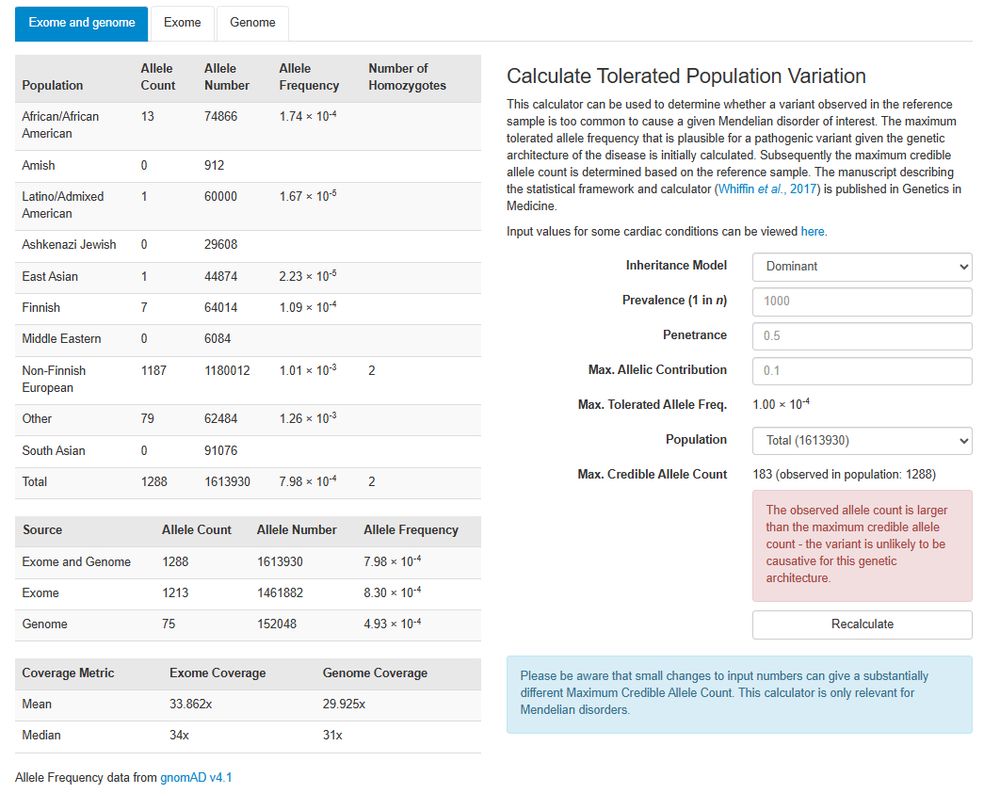
@gnomad-project.bsky.social v4.1 sequence variant data is now displayed; re-annotated using @ensembl.bsky.social Variant Effect Predictor so molecular consequences reflect the gene build on the DECIPHER website. Data will be re-annotated in the future to ensure the annotations remain current
18.06.2025 14:25 — 👍 8 🔁 3 💬 0 📌 0DECIPHER version 11.32 has been released. See the new features at www.deciphergenomics.org #variantinterpretation
18.06.2025 14:25 — 👍 1 🔁 0 💬 0 📌 0Thanks for capturing this and thanks to the workshop organiserat #ESHG2025☺️
DECIPHER is an amazing example of how keeping genomic data patient centred makes it better!
That is all 🎤
@deciphergenomics.bsky.social
(COI see my bio)
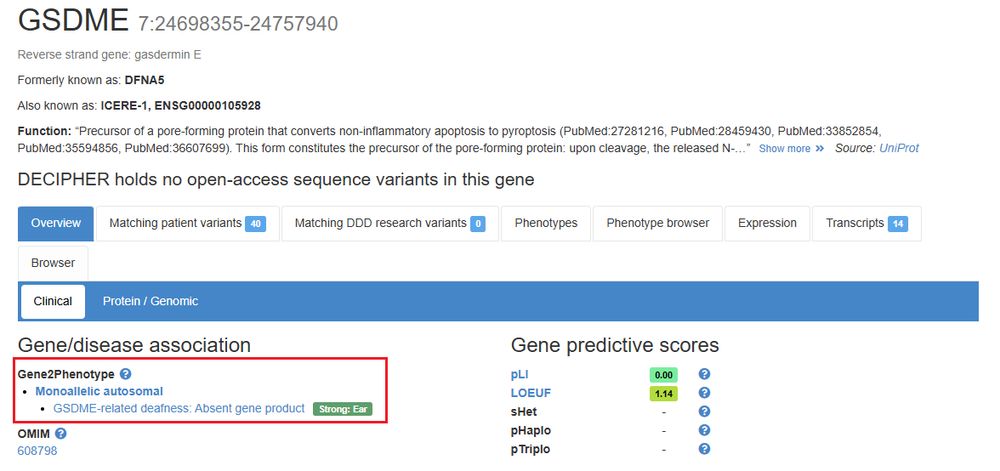
Ear disorders curated by Gene2Phenotype (G2P) are now displayed across the website. This includes curations for 87 genes and 97 Locus-Genotype-Mechanism-Disease-Evidence (LGMDE) threads @gene2phenotype.bsky.social
07.05.2025 13:29 — 👍 5 🔁 1 💬 0 📌 0