The current admissions system based on SES zones and bonus points excludes many kids with A grades and high test scores, especially those from Catholic schools like yours. If we are going to stick with that system then increasing the number of seats seems like the right thing to do to me.
14.05.2025 16:57 — 👍 1 🔁 0 💬 0 📌 0
If this happens, this policy change will prevent discoveries and will result in our children and grandchildren being sicker and dying sooner
07.02.2025 23:48 — 👍 49 🔁 13 💬 2 📌 4
ATTENTION
If you are someone who had an F31-Diversity (or similar) application submitted this cycle, please DM me here, contact me on signal (jeremymberg.78), or email me at jeremymberg@gmail.com.
I will keep all information confidential.
2/n
07.02.2025 16:06 — 👍 195 🔁 270 💬 8 📌 11
If doing CNV calling the default bin size in GATK gCNV for Illumina data is 1kb. I’d look at the histogram and MAD of 1kb bin depths. If its not tight or your regions of interest contain lots of repetitive sequence 5 or 10kb might work (with less resolution)
04.01.2025 14:44 — 👍 2 🔁 0 💬 0 📌 0
Not window size based, but we often use metrics like “Percent bases at 20X coverage” to QC WGS data for variant calling.
04.01.2025 14:24 — 👍 1 🔁 0 💬 1 📌 0
I'm sort of skeptical that that's the case at selective schools, which in my experience liked well-rounded applicants, but maybe things have changed.
11.12.2024 17:45 — 👍 1 🔁 0 💬 0 📌 0
Genuine question -- why not just push for rigorous college-prep level high school humanities classes then? I confess I do not get the appeal of Early College as currently presented. Seems like quasi-vocational "career-track" stuff instead of a chance to actually explore college-level classes early.
11.12.2024 14:59 — 👍 1 🔁 0 💬 1 📌 0
What Wu policies are responsible for the low homicide rate?
11.12.2024 14:49 — 👍 0 🔁 0 💬 0 📌 0

Today is the FINAL day to submit your bids in our Silent Auction for a Healthy Ocean!
Bidding closes at 8 PM Eastern! Jenna and Sarah will be going LIVE on Instagram at 4 PM to showcase some of the incredible auction items—tune in to catch the action! ow.ly/5vVf50UkggC
03.12.2024 19:03 — 👍 2 🔁 3 💬 2 📌 0
I put some #rstats code into Claude and told it to optimize it. It totally rewrote it and got a 50x speed up with identical results. …maybe we actually are cooked?
03.12.2024 21:00 — 👍 4 🔁 0 💬 0 📌 0
Lifelong JP resident. BPS grad and (unhappy) BPS parent.
19.11.2024 14:06 — 👍 1 🔁 0 💬 0 📌 0
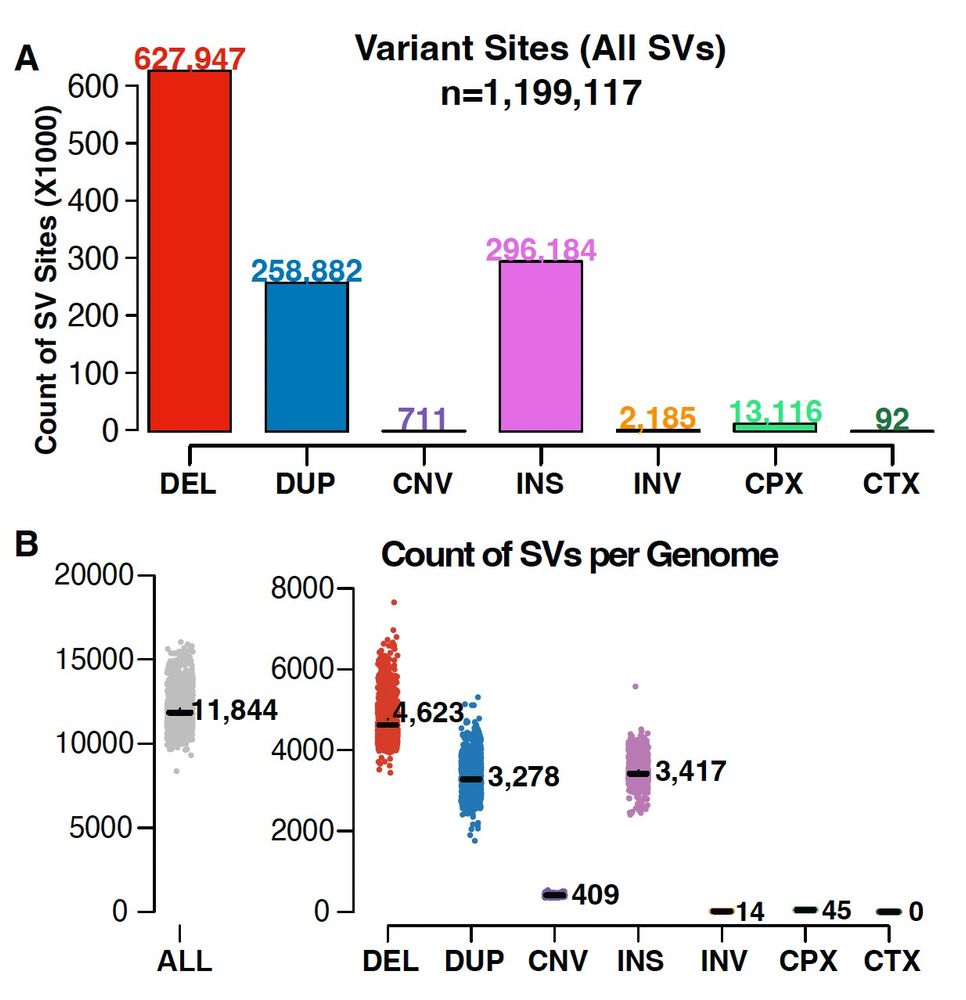
As part of #gnomAD v4, in collaboration with the Talkowski Lab, we have released 1,199,117 genome SVs and 66,903 rare exome CNVs. These data represent the first gnomAD SV dataset released native to the GRCh38 reference genome. (1/2)
02.11.2023 12:58 — 👍 7 🔁 3 💬 1 📌 0
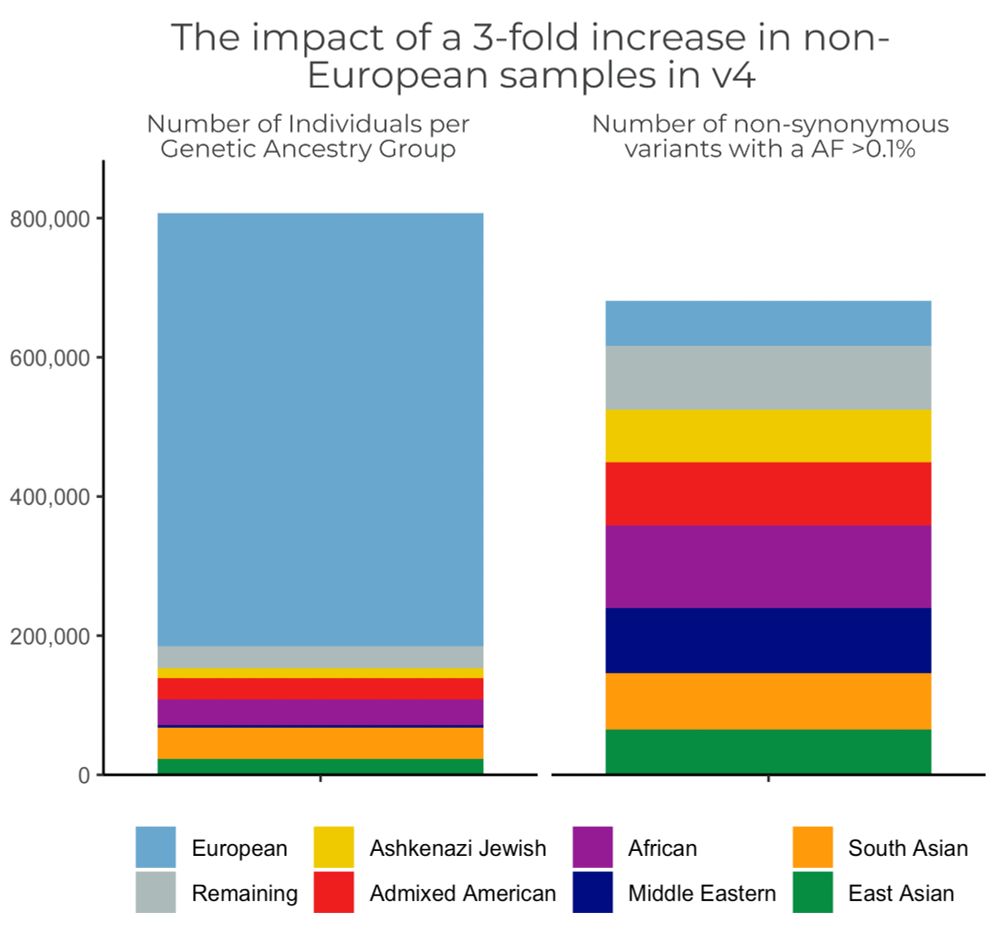
The #gnomAD team is proud to announce the release of gnomAD v4! The v4 dataset includes 730,947 exomes & 76,215 genomes, which is ~5x larger than the v2 & v3 releases combined, & includes nearly 120K indivs of non-European genetic ancestry broad.io/gnomad #ASHG23 (1/11)
01.11.2023 18:35 — 👍 32 🔁 21 💬 2 📌 2
haskell/rust/art/shitposting/assyrian
she/her (like a ship, not like a person)
Editor at large, The Bulwark.
Director, Defending Democracy Together.
Host, Conversations with Bill Kristol.
Never Trump.
Independent AI researcher, creator of datasette.io and llm.datasette.io, building open source tools for data journalism, writing about a lot of stuff at https://simonwillison.net/
I am a pension fund monitor for Fremulon Insurance.
New Book "Big Fan" (with Joe Posnanski) available May 19. Preorder signed copies here: https://josephbeth.com/book/9798217045112
god created him and demanded that he die
assistant professor at ucsf interested in genetics, statistics, etc…
jeffspence.github.io
"A complex decision is like a great river, drawing from its many tributaries the innumerable premises of which it is constituted." - HA Simon
https://adamelkus.com/ + also @ aelkus on twitter
all opinions my own but also your cat's
The Coolidge Corner Theatre is a nonprofit cinema that has been showing the best classic, foreign, independent, & midnight cult films since 1933.
We are a statewide coalition of pro-housing advocates and organizations, standing up for abundant housing for all in communities across Massachusetts.
Bioinformatician, Leader, Founder @fulcrumgenomics.bsky.social
https://www.fulcrumgenomics.com
The GREGoR Consortium (Genomics Research to Elucidate the Genetics of Rare diseases) seeks to develop and apply approaches to discover the cause of currently unexplained rare genetic disorders.
https://gregorconsortium.org/
Good in a crisis and at no other time
First initial last name at gmail dot com
Signal:@KathrynTewson.06
The original #MAPoli newsletter
📪 https://massterlist.com/subscribe/
Programmer. Trying to enjoy arguing on the Internet less. He/Him.
Sometimes post pictures of food or thoughts on books I read.
Economics Faculty @UMassBoston Thinking about cities, housing and labor. Teaching #UrbanEcon. Living in #Dorchester, #Boston. #YIMBY #MAPoli https://kerenhornecon.com
Proud UMass Amherst alum. Recovering politico & current ED of Boston Policy Institute. MA’s Governor is just a weak mayor. Opinions are my own.
The Dorchester Reporter, the leading news source for Boston’s largest neighborhood since 1983.
https://www.dotnews.com/
EPIGENETIC HULK SMASH PUNY GENOME. MAKE GENOME GO. LOCATION: NOT CENTROMERE, THAT FOR SURE


