
Accurate, scalable structural variant genotyping in complex genomes at population scales. #VariantGenotyping #StructuralVariants #PopulationScale #Genomics #Bioinformatics @molbioevol.bsky.social 🧬 🖥️
academic.oup.com/mbe/advance-...
@edg1983.bsky.social

Accurate, scalable structural variant genotyping in complex genomes at population scales. #VariantGenotyping #StructuralVariants #PopulationScale #Genomics #Bioinformatics @molbioevol.bsky.social 🧬 🖥️
academic.oup.com/mbe/advance-...

scTail: precise polyadenylation site detection and its alternative usage analysis from reads 1 preserved 3′ scRNA-seq data. #PolyAsite #scRNAseq #GenomeBiology
genomebiology.biomedcentral.com/articles/10....

Very excited to share new work from my PhD on a new software package for eQTL mapping: quasar. The quasar software package is a C++ program designed to provide a flexible and efficient eQTL mapping. www.medrxiv.org/content/10.1...
22.07.2025 10:15 — 👍 42 🔁 17 💬 2 📌 1
PanVA: a visual analytics tool for pangenomic variant analysis. #Pangenomes #VariantAnalysis #DataVisualization #Genomics #Bioinformatics @biorxiv-bioinfo.bsky.social 🧬 🖥️
www.biorxiv.org/content/10.1...

A Benchmark of Modern Statistical Phasing Methods. #DNAphasing #MethodsBenchmarking #Genomics #Bioinformatics @biorxiv-genomic.bsky.social 🧬 🖥️
www.biorxiv.org/content/10.1...
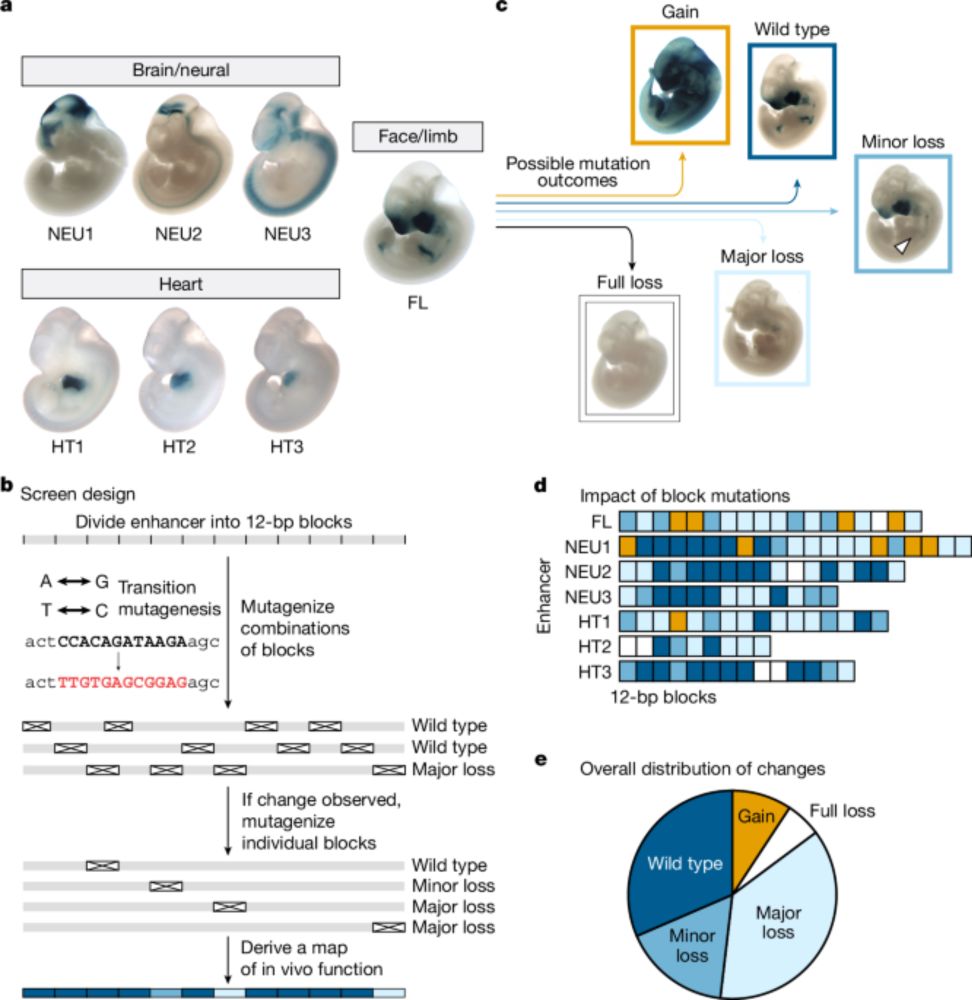
In vivo mapping of mutagenesis sensitivity of human enhancers
www.nature.com/articles/s41...

Preprint on "Improving spliced alignment by modeling splice sites with deep learning". It describes minisplice for modeling splice signals. Minimap2 and miniprot now optionally use the predicted scores to improve spliced alignment.
arxiv.org/abs/2506.12986

cuteFC: regenotyping structural variants through an accurate and efficient force-calling method genomebiology.biomedcentral.com/articles/10...
15.06.2025 14:15 — 👍 3 🔁 1 💬 0 📌 0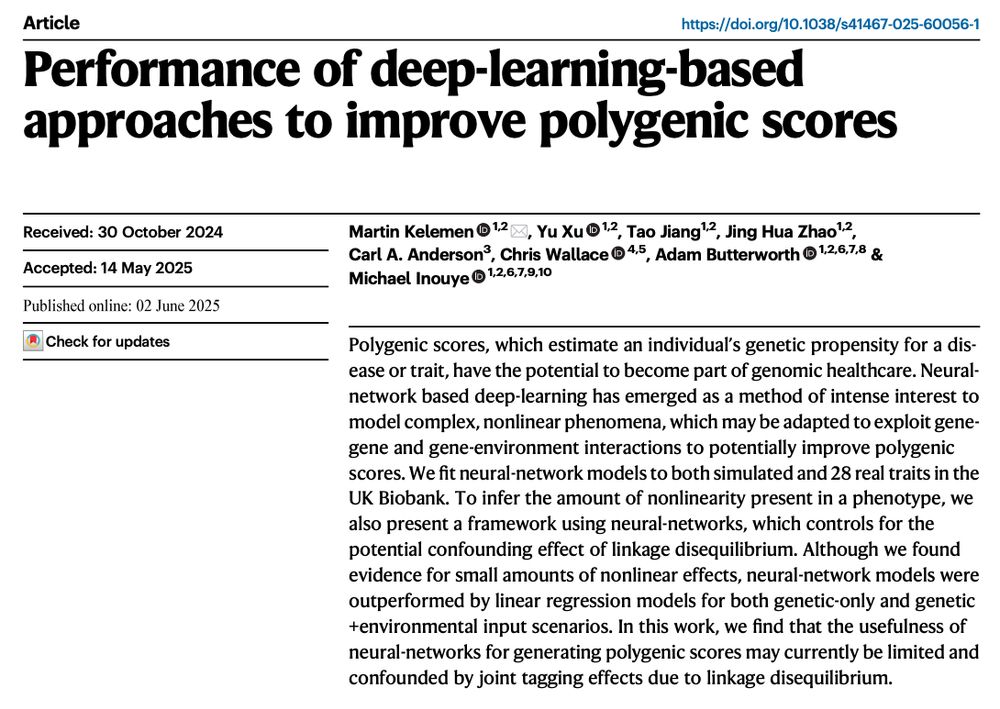
📣 Latest from the lab: Performance of deep-learning-based approaches to improve polygenic scores www.nature.com/articles/s41...
Its thought deep learning will substantially improve PGS but the reality is MANY have tried but no/little gain has been seen so far. Here we report our negative results.


SNP calling, haplotype phasing and allele-specific analysis with long RNA-seq reads www.biorxiv.org/content/10.1... 🧬🖥️🧪
longcallR: github.com/huangnengCSU...
Nextflow workflow: github.com/huangnengCSU...

Check out our latest blog post to discover how to build custom Docker images for Seqera Studios, with real-world examples including Marimo, Streamlit, CELLxGENE, and Shiny! 🌟
📖 Read the blog post: hubs.la/Q03psL-t0

Happy to share our new study on genetic & environmental contributors to age-related decline in ~100K UK Biobank participants!
Here, we used simulation work + longitudinal GWAS and downstream analyses to explore risks involved in cognitive/physical decline
(1/)🧵🧵
shorturl.at/99gqL

ConsensuSV-ONT: A modern method for accurate structural variant calling www.nature.com/articles/s41... 🧬🖥️🧪 Nextflow: github.com/SFGLab/Conse...
19.05.2025 15:31 — 👍 4 🔁 2 💬 0 📌 0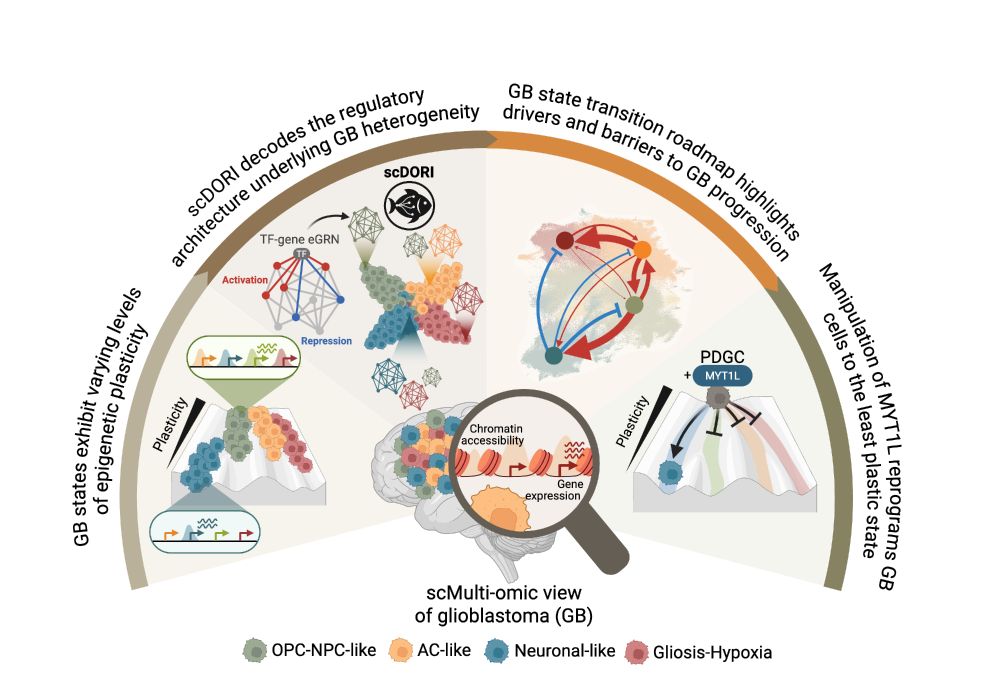
🧠 Excited to share my main PhD project! We mapped the regulatory rules governing Glioblastoma plasticity using single-cell multi-omics and deep learning. This work is part of a two-paper series with @bayraktarlab.bsky.social @oliverstegle.bsky.social and @moritzmall.bsky.social, Preprint at end🧵👇
16.05.2025 10:04 — 👍 76 🔁 29 💬 1 📌 6
📣 Mark your calendars! The 2025 edition of the scverse conference will take place on 17-19 November at Stanford University (US) scverse.org/conference20...
Call for abstracts and registrations coming soon!

Our work looking at the effects of using autosomal assumptions for calling variation on X and Y. We used simulated data to compare to a ground truth.
Tldr: Accurate ploidy is important for reducing false positives; appropriate alignment reduces false negatives.
www.biorxiv.org/content/10.1...


py_ped_sim: a flexible forward pedigree and genetic simulator for complex family pedigree analysis bmcbioinformatics.biomedcentral.com/articles/10.... 🧬🖥️🧪 github.com/MiguelGuarda...
08.05.2025 18:30 — 👍 4 🔁 3 💬 0 📌 0
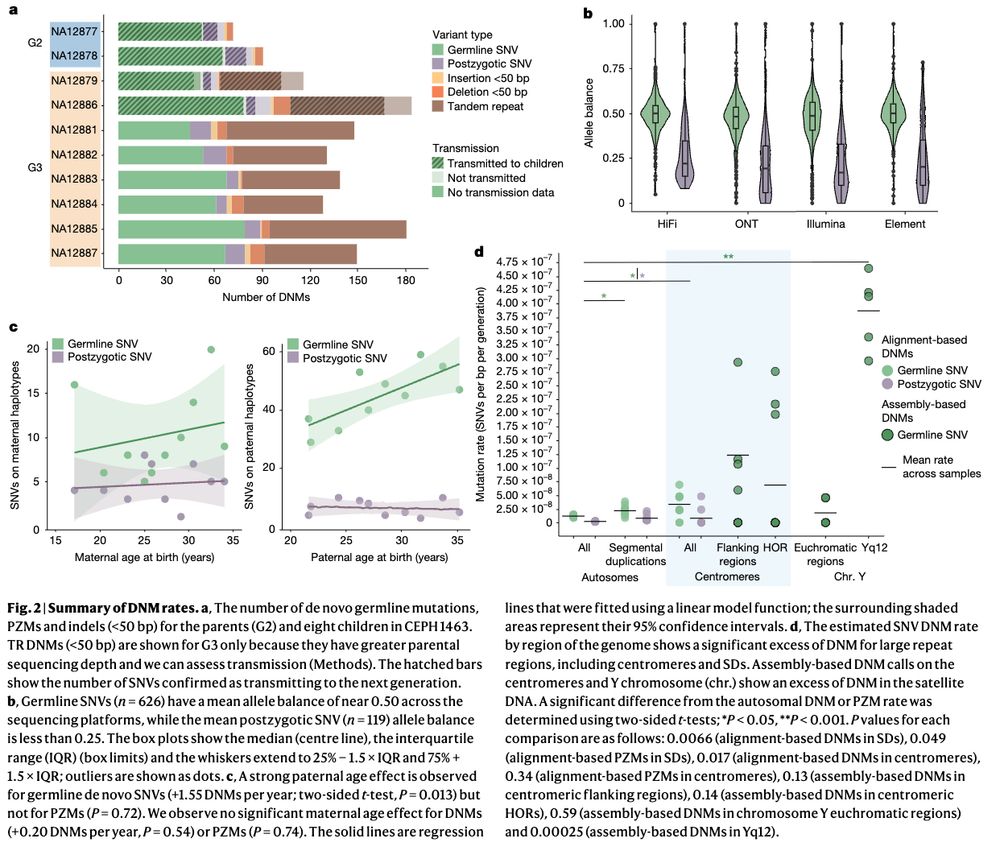


Human de novo mutation rates from a four-generation pedigree reference www.nature.com/articles/s41... 🧬🖥️🧪
02.05.2025 18:30 — 👍 17 🔁 6 💬 0 📌 1
(1/3)🧬 Tissue Genetics: Prevalent genetic effects extend beyond cell boundaries, shaping gut tissue immunology. Work with Aviv Regev, Ramnik Xavier @broadinstitute.org, Kirk Gosik, Heping Xu, Gary Churchill, Kushal K. Dey and other great collaborators.
www.biorxiv.org/content/10.1...
Big updates for the Nextflow @vscode.dev extension! 🚀
🔍 Workflow view – Visualize logical structure of pipelines
⚙️ Process view – See all processes in one place
🌐 Seqera view – Manage connected CEs directly in the IDE
📚 Resources view – Quickly access Copilot, AI tools & training

Some encouraging news for cross-gene generalization of allele effects in S2F models. www.biorxiv.org/content/10.1...
16.04.2025 01:46 — 👍 15 🔁 7 💬 1 📌 0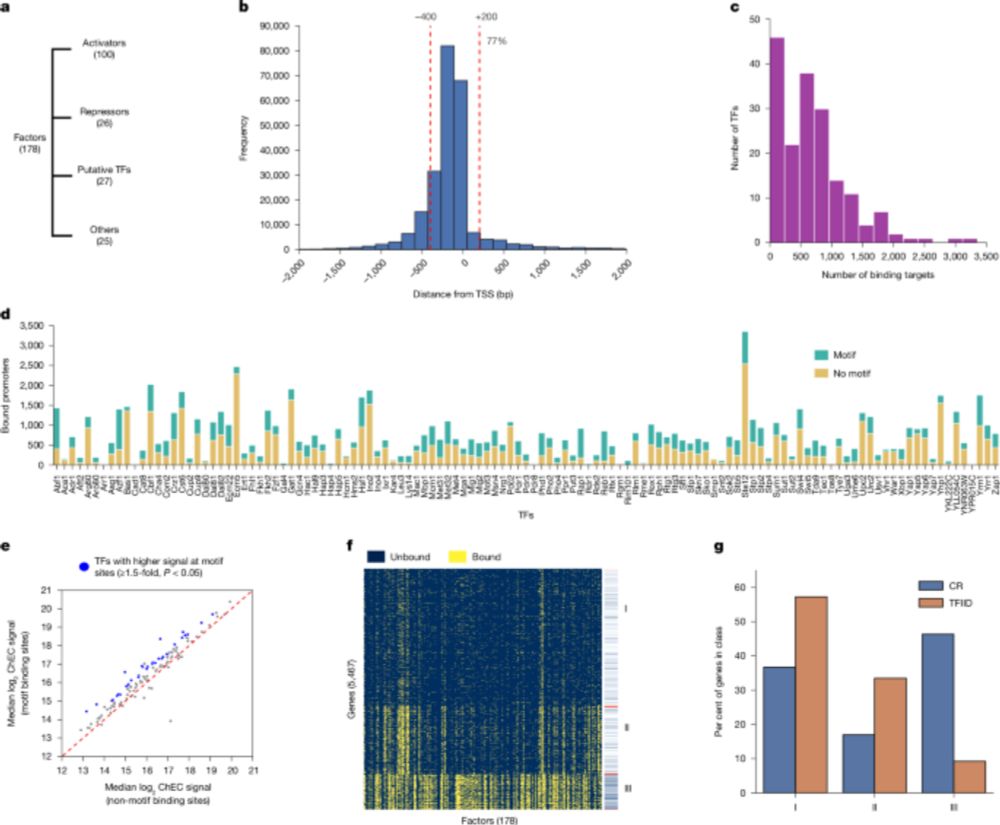
To the top of the "to-read" list. Looks like a heroic amount of work from the Hahn lab (large-scale ChEC-seq compendium!) www.nature.com/articles/s41...
16.04.2025 16:04 — 👍 56 🔁 28 💬 3 📌 2
New paper out in Genome Biology! 🎉
We lay out best-practice guidelines for releasing variant effect predictors, developed through the Atlas of Variant Effects Alliance @varianteffect.bsky.social
Open, interpretable, and clinically useful VEPs are the goal.
📄 doi.org/10.1186/s130...

🎙️Just released: Episode 51 of the @nextflow.io podcast!
@ewels.bsky.social and Ben Sherman discuss the upcoming #Nextflow strict syntax - cleaner code, better errors, and a more consistent language framework.
Learn what's changing and how to prepare your pipelines: hubs.la/Q03hpNvx0

scnanoseq nf-co.re/scrnaseq: an nf-core [Nextflow] pipeline for Oxford Nanopore single-cell RNA-sequencing www.biorxiv.org/content/10.1... 🧬🖥️🧪 github.com/nf-core/scna...
11.04.2025 20:00 — 👍 26 🔁 12 💬 0 📌 1distQTL: Distribution Quantitative Trait Loci Identification by Population-Scale Single-Cell Data https://www.biorxiv.org/content/10.1101/2025.04.04.647121v1
10.04.2025 22:33 — 👍 5 🔁 4 💬 0 📌 0We are excited to announce that Studios is now generally available!🎉
Studios lets you easily transition from bioinformatics workflows to secure, interactive analysis environments on your own infrastructure.
💡Read the blog to find out more: hubs.la/Q03gy11J0

🚀 Truvari v5.0 is here! 🎉
What’s new?
🔹 Enhanced symbolic variant support for <DEL>, <DUP>, <INV>
🔹 Robust BND comparison for cross-representation SV matching
🔹 Improved SV sequence similarity & HUGE SV support
🔹 Cleaner UI & Revamped API
👉 More: github.com/ACEnglish/tr...
#Genomics #Bioinformatics

Great to see that sawfish, our new HiFi SV caller, is accepted for publication in Bioinformatics! Sawfish emphasizes local haplotype modeling to improve SV representation and genotyping in both single and joint-sample analysis. Advance-access article now available: (1/n)
doi.org/10.1093/bioi...