The UKB-PPP consortium should receive the first tranche of ~100k samples this autumn and subsequent tranches throughout 2026. Data releases to all approved UKB researchers will begin next year.
11.01.2025 14:32 — 👍 1 🔁 0 💬 1 📌 0
*Alden Scientific, Amgen/deCODE genetics, AstraZeneca, Bristol Myers Squibb, Calico Life Sciences, Genentech, GSK, Isomorphic Labs, Johnson & Johnson, Merck Group, Novo Nordisk, Pfizer, Regeneron and Takeda.
10.01.2025 08:18 — 👍 1 🔁 0 💬 0 📌 0
I'm deeply grateful to the hundreds of collaborators working across fourteen biopharmaceutical companies*, two technology vendors, and one remarkable biobank for making this possible.
10.01.2025 08:18 — 👍 1 🔁 0 💬 2 📌 0
This initiative again underpins the transformative value of public-private partnerships for drug development and healthcare and reemphasizes the quiet 'revolution' happening in the field of proteomics.
10.01.2025 08:18 — 👍 1 🔁 0 💬 1 📌 0
This will allow us to build foundational AI models for biological discovery, study biomarkers that change over time in response to changes in health states, identify surrogate blood biomarkers for MRI endpoints, and - ultimately - develop better medicines & diagnostics.
10.01.2025 08:18 — 👍 1 🔁 0 💬 1 📌 0
We will begin by profiling 300k samples, comprising approximately 250k samples collected at baseline visits and 50k samples taken at various follow-up assessments. We aim to incorporate an additional 250k baseline samples and 50k repeat samples pending additional funding.
10.01.2025 08:18 — 👍 1 🔁 0 💬 1 📌 0

Launch of world’s most significant protein study set to usher in new understanding for medicine
I'm delighted to announce that the Pharma Proteomics Project will commence full-scale proteomic profiling of the UK Biobank in 2025. We have selected the Olink Proteomics Explore HT platform and Ultima Genomics UG 100 sequencers for this unprecedented study.
www.ukbiobank.ac.uk/learn-more-a...
10.01.2025 08:18 — 👍 40 🔁 16 💬 1 📌 3
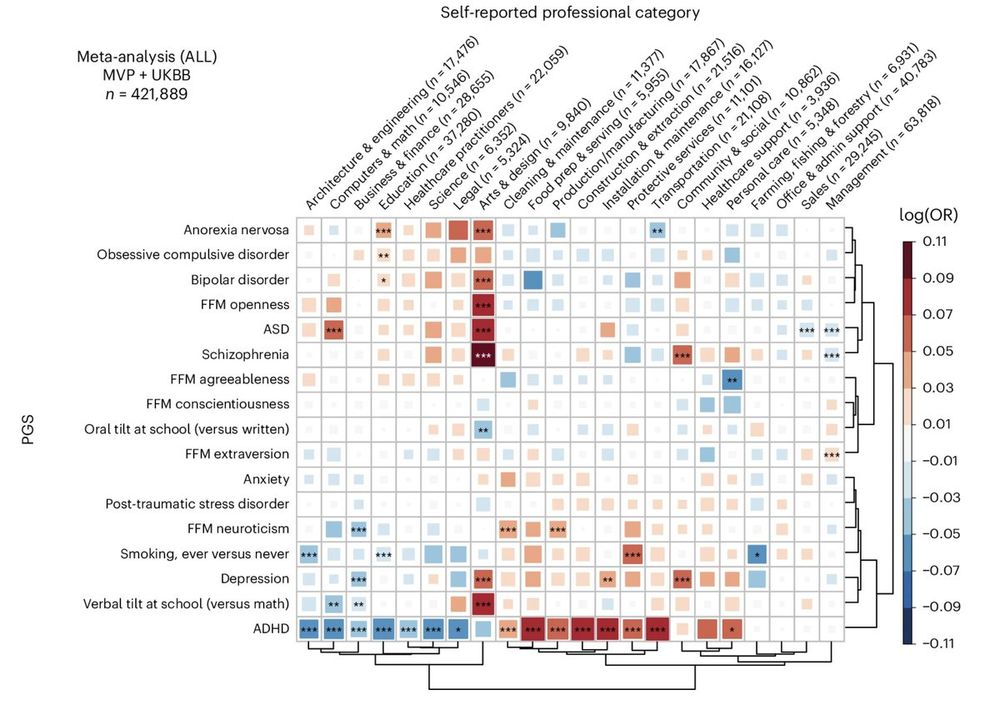
Association of polygenic scores for neuropsychiatric traits with self-reported professions based on analysis of 420k individuals from UK Biobank and Million Veteran Program. Look at the 'arts & design' category. Artistic talent comes at a cost--a piece of your mind :)
12.12.2024 06:14 — 👍 118 🔁 40 💬 7 📌 16
Possibly, but Alamar is low-plex by design (n~250) & captures many low-abundance proteins - not necessarily the most widely studied. Mike’s comment & Jochen’s review offer important perspectives; abundance levels seem to influence confidence in cross-platform detection (higher = more confidence).
21.11.2024 20:36 — 👍 1 🔁 0 💬 0 📌 0
Yes, “good” is relative to what’s been reported before. It’s the median correlation across 217 commonly measured proteins, with a wide range overall. More documentation of the antibodies used +/ epitopes bound could partially address the problem, but might be impractical to implement.
21.11.2024 08:04 — 👍 5 🔁 0 💬 2 📌 0
We’ve found Olink & Alamar to correlate quite well (~60%) overall. Maybe unsurprising, since both are antibody-based.
Lack of correlation b/w SomaScan & others might reveal interesting facets of the aptamer tech (protein folding, PTMs, etc) but 100% agree that infighting is bad for the field.
20.11.2024 23:35 — 👍 7 🔁 0 💬 1 📌 1
Science journalist covering all fields. Formerly an editor at New Scientist and Nature. Particular fan of health, exercise, mushrooms, amphibians and marine life 🧪🐸 🍄
Selection of articles here: https://www.newscientist.com/author/chris-simms/
A leading life science journal that champions high-impact research across all disciplines—from molecules to ecosystems. We offer innovative formats and collaborative editorial support to ensure your work achieves its full scientific impact.
Strategy & Partnering @Metabolon
Multiomics | Science | Outdoor life
#metabolomics
https://www.linkedin.com/in/timdelany/
Senior Editor at @plosbiology.org; Scientist, Humanist, Optimist and Recovering Academic. All opinions my own.
PhD candidate in Cell & Molecular Biology (UofA) studying the genetics behind stress responses of wild yeast
Senior Manager @ Oracle Health. Interested in clinical LLMs, ML + genetics!! Ex-insitro, Microsoft AI4Good, UW. Biriyani expert. Kolkata-> Seattle. Views are my own.
Scholar: https://shorturl.at/QTBGR
Linkedin: https://shorturl.at/WuGl
Physician-scientist, Adult genetics, Human genetics research, Baylor College of Medicine. Thoughts expressed = my own.
Medicine, Genetics, Statistics, Kidney Disease, Columbia University. My posts represent my own views and not those of Columbia University.
Physician Scientist | Nephrology | Translational Research | Single-Cell & Spatial Omics | Precision Medicine | Kidney Innovation | Advocate for Patient-Centered Science | Views My Own
Passionate about #GWAS #Metabolomics #Proteomics #Glycomics #Epigenomics, Professor @WeillCornell
Medicine - Qatar, Blog on http://metabolomix.com, http://suhre.fr
Human Genetics for Drug Discovery
Inter-dimensional cancer biologist. Director of the @genomescience.bsky.social at @umbaltimore.bsky.social.
Public Health #Bioinformatician. Wants to sequence ALL THE THINGS. Personal account with alternative spellings and grammar structures. She/her
Computational evolutionary biologist. Associate Professor at @sandiegostate.bsky.social, athlete, insomniac, writer, singer, polyglot, polymath, Out100 honoree. Open science advocate. He/him. 🏳️🌈🇮🇳🇺🇸🐞🌱🐢🧬🏋🏾♂️🎤✍🏾 www.sethuramanlab.com Views are mine and mine alone.
Avian influenza & avian virome ecology | "Expert in Ducks" | (she/her) | michellewille.com
Postdoc. Omics of reproduction, development, cancer, and conflict.
Opinions are my own.
https://seantbresnahan.com/assets/docs/CV_STB_122925.pdf
https://bhattacharya-lab.com