It is my great honor to join the amazing NGL cohort at
@alleninstitute.bsky.social. I am very much looking forward to connecting and collaborating with everyone in the future!
19.12.2024 16:37 — 👍 4 🔁 1 💬 0 📌 0
#FirstPost
Happy to announce that our research group is moving from UCLA to UPenn in the Dept of Genetics and Institute of Biomedical Informatics at Perelman School of Medicine!
So thankful to the amazing UCLA community for 12 wonderful years! #BruinForever
Excited for new adventures in Philly!
28.11.2024 15:07 — 👍 85 🔁 5 💬 10 📌 1
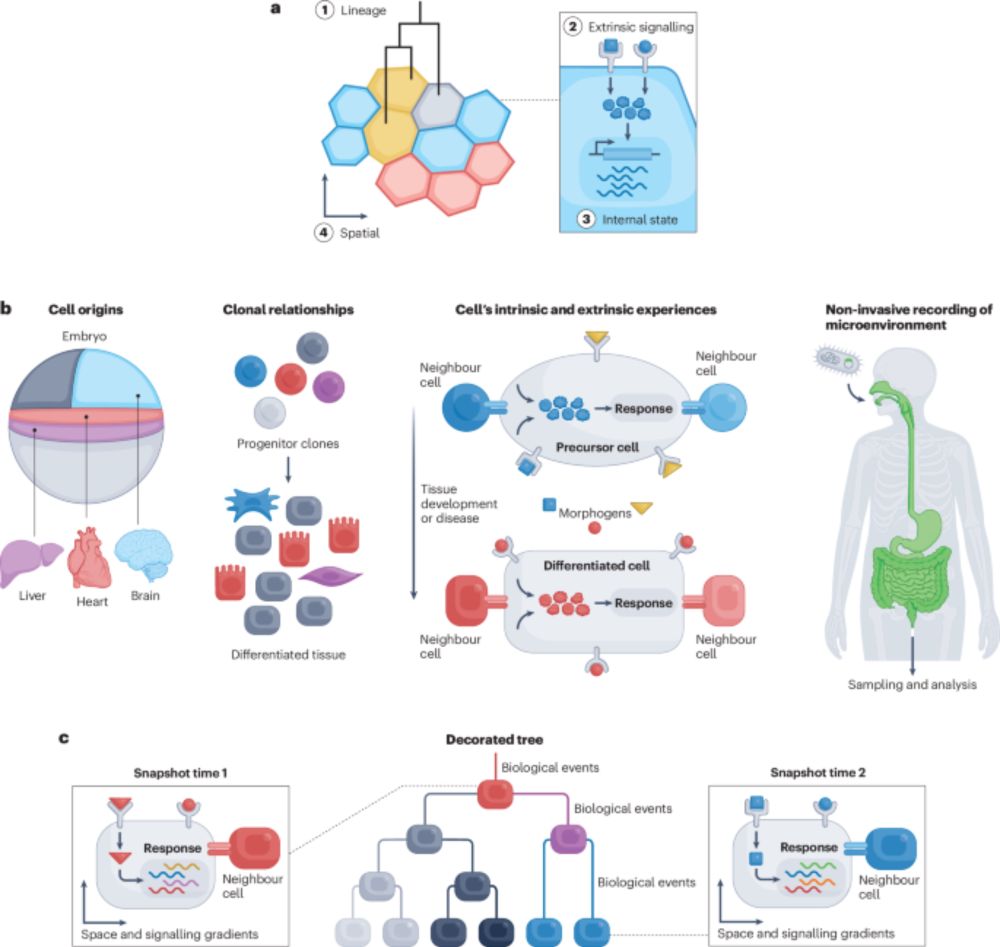
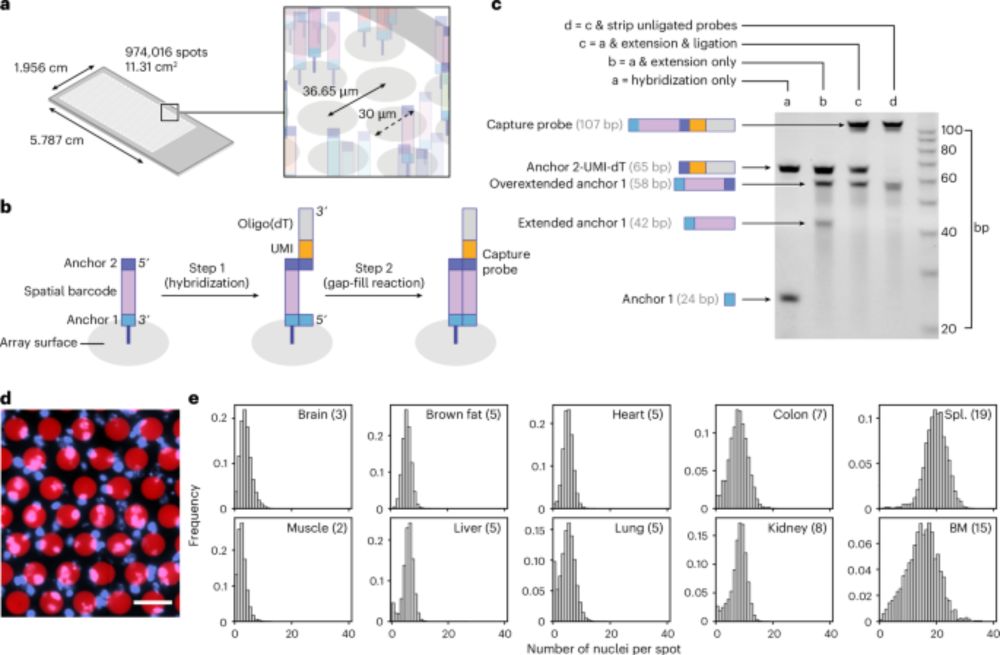
The lives of cells, recorded - Nature Reviews Genetics
Recent advances in genome engineering are enabling the recording of cellular histories into genomes, with single-cell and spatial omics technologies enabling their reconstruction into cellular lineage...
"The lives of cells, recorded"--our new review on genomic recording systems and how they can reveal the dynamics of multicellular development. A pleasure to work on this with amazing colleagues from the Allen Discovery Center for Cell Lineage Tracing.
www.nature.com/articles/s41...
25.11.2024 17:28 — 👍 144 🔁 58 💬 2 📌 4
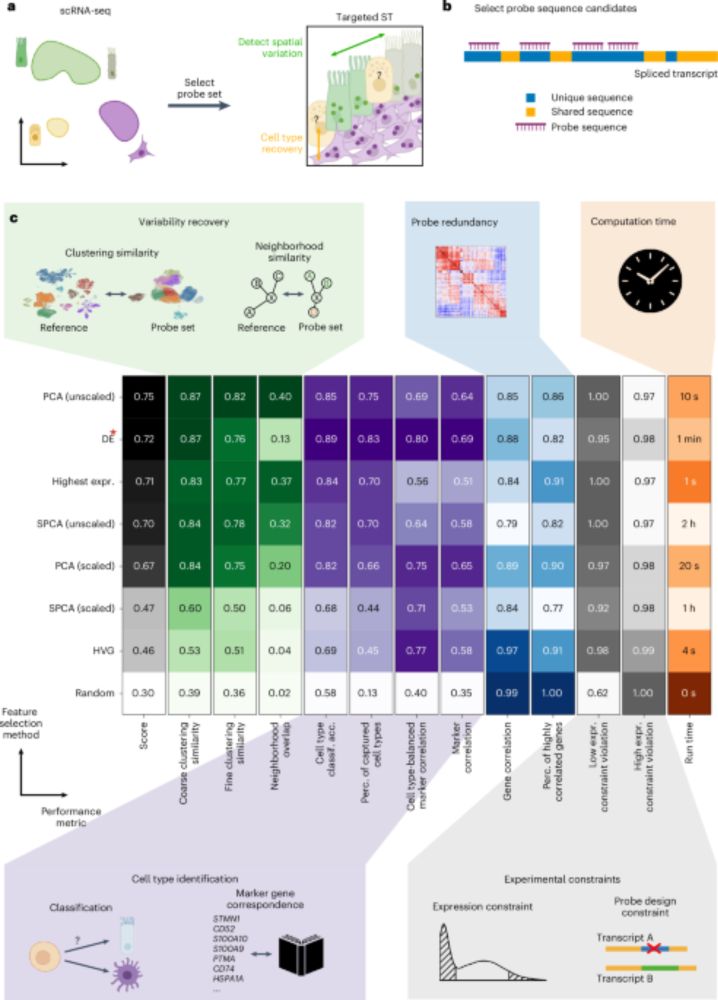
Probe set selection for targeted spatial transcriptomics - Nature Methods
Spapros is a probe set selection pipeline for targeted spatial transcriptomics that optimizes for both transcriptional and within-cell type variation.
Targeted ST pre-selects 100s cell type marker genes. This can exaggerate diff between types while lacks continuity within types. Spapros - probe design pipeline - selects the minimal set of genes to recover both cell type identification and within-type variation.
www.nature.com/articles/s41...
25.11.2024 14:48 — 👍 1 🔁 0 💬 0 📌 0

I will be starting my lab at Stanford in 2025. We are looking for talented postdocs / grad students to join us to develop single cell sequencing, spatial genomics, machine learning, functional genomics, live cell imaging tools to understand the cell communication across space and time. Please RT!Thx
25.11.2024 14:44 — 👍 3 🔁 1 💬 0 📌 0