
Most genetic interaction studies measure gene relationships in one condition, but environment can reshape which genes depend on each other.
Today in Molecular Cell, Luke Gilbert & team deliver the first systematic, multi-condition map of genetic rewiring in human cells.
19.02.2026 17:36 — 👍 13 🔁 6 💬 1 📌 0
Happy to share our new pre-print on the dependency of ALT-positive tumors on the DNA translocase SMARCAL1. Outstanding work of @angelotaglialatela.bsky.social in collaboration with the Min, Lazzerini Denchi and @cejkalab.bsky.social laboratories! www.biorxiv.org/content/10.6...
17.02.2026 03:57 — 👍 20 🔁 9 💬 0 📌 0

A group of about 30 people pose together in a bright office space.
Today, we’re launching a new company, ALTx Therapeutics. Founded by Simon Boulton, the company will develop the first precision therapies targeting ALT cancers, in partnership with Slingshot Therapeutics Limited and @crhorizons.bsky.social.
Find out more ➡️ www.crick.ac.uk/news/2026-02...
05.02.2026 12:45 — 👍 20 🔁 7 💬 0 📌 0

A 1-day Melbourne workshop on Gene Editing in Blood and Immune Cells. February 20, register at editblood.com
18.01.2026 22:43 — 👍 5 🔁 0 💬 0 📌 0

2/Led by @hildapickett.bsky.social, with co-investigators @genomestability.bsky.social, @lisannes.bsky.social, Jacob Lewis, @yuhenglau.bsky.social, Sarah Henrikus, & the Cesare Lab, this project brings together expertise in telomere biology, DNA repair, biochemistry, drug discovery, & cell biology.
12.01.2026 01:50 — 👍 4 🔁 1 💬 1 📌 0
1/Delighted to be part of a team from @cmri.bsky.social, @sydney.edu.au, the University of Wollongong, and SVI (Melbourne) awarded a $5M NHMRC Synergy Grant to explore: “A mechanistic approach to developing precision therapies for ALT-dependent cancers.”
www.nhmrc.gov.au/funding/find...
12.01.2026 01:50 — 👍 19 🔁 4 💬 3 📌 0

Just updated my X/Twitter profile. I suggest you try to encourage any colleagues remaining on that pit to do the same. Such a shame, as it was such an important place of science until not that long ago. :-(
12.01.2026 00:55 — 👍 3 🔁 0 💬 0 📌 0
8–11 June 2026 | 📍 Heraklion, Crete 🇬🇷
Get ready for Machines on Genes 2026, the 94th Harden Conference by the @biochemsoc.bsky.social — four days of molecular mechanisms and friendly discussions! 🧬☀️🔬❄️
Organised by @lapassmore.bsky.social, Dana Branzei, me.
09.01.2026 10:14 — 👍 15 🔁 11 💬 1 📌 0
Yep, me too. I have also been getting this error quite a lot in the past couple weeks: "Model inference failed". Usual wait times are 2hr...
17.12.2025 04:04 — 👍 1 🔁 0 💬 0 📌 0

No, not that FANCL!
New paper on FancL mutant mouse just out in @bloodadvances.bsky.social shows essential function of E3 RING ligase activity to phenotypes of #FanconiAnemia plus, Black Friday deal: a new model for testing gene editing therapies: ashpublications.org/bloodadvance...
27.11.2025 02:03 — 👍 9 🔁 1 💬 0 📌 0
Chromatin fatigue: DNA repair alters the chromatin environment and introduces heritable variation in gene expression in a larger region around the lesion! Amazing achievement by @sbantele.bsky.social and Jiri Lukas published in @science.org 🙌 Happy we could contribute. See 👇
12.11.2025 03:55 — 👍 38 🔁 10 💬 0 📌 0

Registration is open!🧬
Join us at Egmond aan Zee (April 19–24, 2026) for the next DNA Repair Meeting. We have an amazing line-up of speakers.
📅 Deadline for early registration: Jan 10, 2026
🔗 dnarepairmeeting-egmond2026.com
12.11.2025 15:04 — 👍 22 🔁 13 💬 1 📌 2

DNA REPAIR/GENOME STABILITY CONFERENCES
🧬 List of 2026 DNA repair and genome stability conferences, now updated. Please let me know if there is anything missing!
docs.google.com/spreadsheets...
09.11.2025 22:33 — 👍 23 🔁 10 💬 0 📌 0
 31.10.2025 01:59 — 👍 0 🔁 0 💬 0 📌 0
31.10.2025 01:59 — 👍 0 🔁 0 💬 0 📌 0

I attended the Australian Cell Cycle, DNA Repair and Telomere Meeting for the first time, and now I wish I could have attended it earlier.
Thanks much to the organisers and especially to @genomestability.bsky.social for putting such a nice meeting together.
I am looking forward to the future ones!
27.10.2025 01:27 — 👍 6 🔁 2 💬 0 📌 0

It was a great honour to present the AGCT (Australian Genome instability, Cell cycle and Telomere) award to Prof Kum Kum Khanna yesterday evening at #ADCTM25. Kum Kum has had an amazing career in discoveries relating to these fields in Australia, and been a remarkable leader and mentor to many.
22.10.2025 04:38 — 👍 10 🔁 1 💬 1 📌 0


Poster session 1 was such a hive of activity and discussion at Melbourne museum today! #ACDTM25
20.10.2025 18:14 — 👍 3 🔁 2 💬 0 📌 0

Its so awesome to have Alan D'Andrea here in Melbourne today for #ACDTM25. What a great talk framed by his classic work on FA!
20.10.2025 18:09 — 👍 8 🔁 0 💬 0 📌 0

Ever wondered what S-phase tastes like? 3 lucky presenter prize winners will find out (+Amazon voucher!) at #ACDTM25. You can still register for this fantastic Melbourne meeting Oct 20-22. Official 10 year old taste tester: "lemon excision repair" is the best flavour www.australiancellcycle.org
13.10.2025 02:25 — 👍 10 🔁 4 💬 0 📌 0
Australian Cell Cycle, DNA repair and Telomere meeting draft schedule is now online!
Join us with 5 plenary speakers, 16 invited national speakers, 22 selected speakers, 61 poster presenters and lots and lots of fun science. Less than one month to go.
australiancellcycle.org/draft-schedu...
25.09.2025 01:47 — 👍 9 🔁 3 💬 1 📌 1
Group Leader, MRC Laboratory of Molecular Biology & Joint Head of the Division of Protein & Nucleic Acid Chemistry. Senior Executive Editor, Nucleic Acids Research.
Research Fellow at the University of Birmingham, investigating the Alternative Lengthening of Telomeres mechanism. Supported by an MRC Career Development Award.
https://www.dundeegenomeintegrity.org
https://www.dundeegenomeintegrity.org/researcher/agostina-bertolin/
Understanding genomic stability and human disease through a DNA replication lens| Wellcome-funded Group Leader| Former Diffley postdoc, Crick
Postdoctoral researcher at the Mailand lab. Interested in histone transcription, but also follow any climbing-related content
Postdoc fellow in the @gerlichlab.bsky.social • Genomic integrity and nuclear processes • He/him
https://orcid.org/0000-0003-0953-8010
Molecular Biologist (aemonten.github.io) 丨Chief Editor @ CSH Protocols (cshprotocols.cshlp.org) 丨Head of the Integrity in Publishing Group at CSHL Press 丨Chair "Molecular Biosystems Conference" (molbiosystems.com) 丨(Oxford) Comma King 丨Central Dogma Police
PhD Candidate in the Zhao Lab at MSKCC/WCGS interested in genomic integrity and SUMO 🧬
Curious about genome stability,currently studying how Arabidopsis keeps it together.
PhD student at Institute of Experimental Biology, and Masaryk University, Czech Republic
building a new 🧠 in the skull of the old [ADHD enjoyer] ~ wriggle 'n twist like ⚡ to the ground, freedom is an endless 🔥 [🏴] ~ vulgar pseud [call center worker] ~ confused by nonhuman ethics & agency; studying biophysics & geometry 101 [🆘️]
Columbia University & HHMI
We study principles of biological time control with a focus on the emerging concept of autonomous clocks. See more at: aydoganlab.com
Team leader exploring DNA replication events using genetics and genomics at IMB Mainz
nucleic acids, polymerases, helicases, and mitochondria at Tufts University
HHMI Hanna Gray Fellow and Assistant Professor at @CornellMBG studying the evolution and molecular mechanisms of chromosome segregation in fungi 🍄| PhD @ScienceStowers | she/her | 🇲🇽
Scientist interested in PARPs, PTMs and genome stability
Structural biologist working to understand DNA replication at the MRC-LMB
Researcher at Heidelberg University Hospital
ICMUB (institut de chimie moléculaire de l'Université de Bourgogne)
Université Bourgogne Europe / CNRS / Dijon, France
https://icmub.ube.fr/
Royal society research fellow and lecturer at the University of Sheffield. Working on DNA damage and repair in mitosis. All views are my own :)
Professor, PI, Cell and Molecular Biologist. Study genome integrity and cancer etiology. Fun of Xenopus laevis. views=own
Postdoc Cimprich lab, Duxin lab PhD alumna







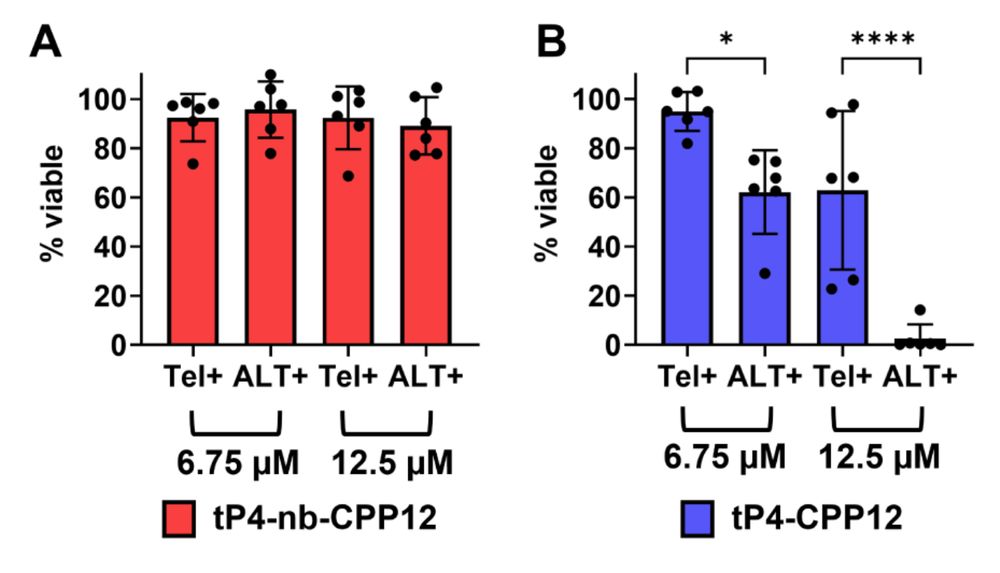







 31.10.2025 01:59 — 👍 0 🔁 0 💬 0 📌 0
31.10.2025 01:59 — 👍 0 🔁 0 💬 0 📌 0










