
We just released version v0.3.1 of 🍦Icecream!
Importantly, this fixes a bug in missing wedge correction. Please update, and let us know how it works for you!
#CryoET #TeamTomo
github.com/swing-resear...
@dhrebik.bsky.social
Postdoc @JohnBriggsGroup @MaxPlanck Institute of Biochemistry in Martinsried | PhD from PlevkaLab | #StructuralBiology of #Viruses #HIV | #CryoEM | #Memes Twitter/X: @F4ustus

We just released version v0.3.1 of 🍦Icecream!
Importantly, this fixes a bug in missing wedge correction. Please update, and let us know how it works for you!
#CryoET #TeamTomo
github.com/swing-resear...
It’s been a while since I put together the Structural Virology starter pack, and I’m sure some new folks have joined Bluesky since then. There’s still plenty of space to fill, so if you’d like to be added to the list just let me know.
#virology #cryoEM
go.bsky.app/Qxv95BL
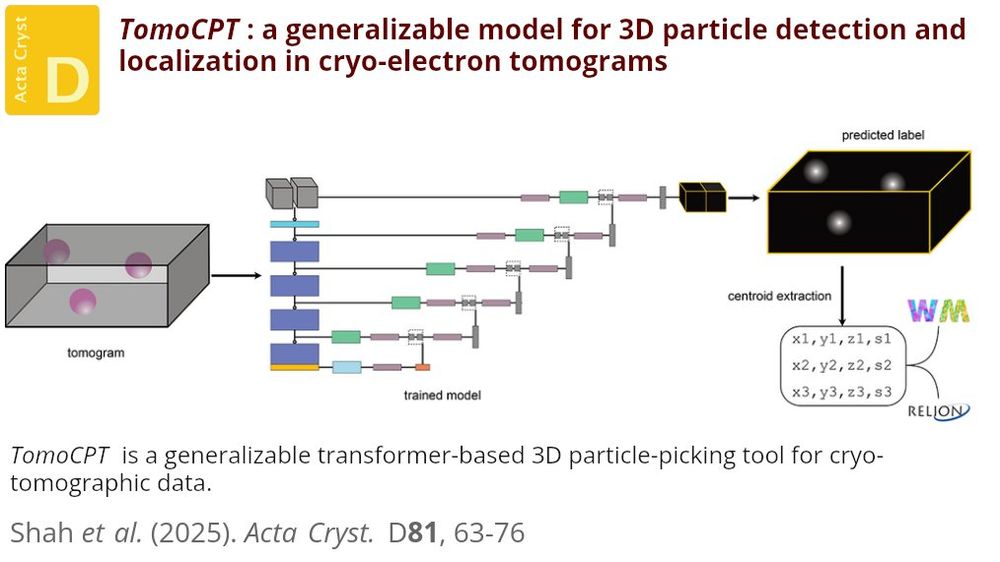
TomoCPT is a transformer-based solution that reformulates particle detection in cryo-ET as a centroid-prediction task using Gaussian labels #CryoET #ParticlePicking #SubTomogramAveraging doi.org/10.1107/S205...
05.03.2025 14:47 — 👍 8 🔁 3 💬 1 📌 0Visually, the cell contains a surprisingly low amount of ribosomes when I compare it to tomograms from other organisms I have seen before (bacteria, eukaryota). Is that a known feature of Archaea?
08.11.2025 20:35 — 👍 4 🔁 0 💬 1 📌 0
Excited to launch an openRxiv partnership with the scientist-run AI review service qed (@qedscience.bsky.social), the brainchild of @odedrechavi.bsky.social 1/n
openrxiv.org/enabling-rev...

An Asgard archaeon with internal membrane compartments
Brilliant study led by @fmacleod.bsky.social and Andriko von Kügelgen. Tight collaboration with @buzzbaum.bsky.social and lab. Congrats to all authors!
www.biorxiv.org/content/10.1...

Glad to see our antibody design paper finally out in
@nature.com
(and congrats to lead authors and everyone involved )!
At Xaira we are excited about pushing antibody design further to bind harder targets and make drugs for unmet medical needs.
Paper:
www.nature.com/articles/s41...
At some point, Doppio will replace relion's gui. Already you can do more in Doppio than in relion! 🤩
04.11.2025 18:15 — 👍 22 🔁 4 💬 0 📌 1



Celebrate #NationalCatDay with the cats of RCSB PDB!
29.10.2025 16:55 — 👍 30 🔁 5 💬 0 📌 0We are looking for a computational postdoc to work with us on new optimisation algorithms to make #RELION even better. Join our bubbly team at the @mrclmb.bsky.social in Cambridge, UK. 🤗 RTs appreciated.
mrc.tal.net/vx/appcentre...
Measurement of atomic scattering factors by cryo-electron microscopy www.biorxiv.org/content/10.1101/2025.10.24.683059v1 #cryoEM
25.10.2025 10:03 — 👍 11 🔁 4 💬 0 📌 0
A Review BMC Biology from the Mechanobiology Collection introduces the fundamental concepts of curvature, discusses its effect on cellular processes and behaviors at the cell-scale, and offers insights into emerging perspectives on the roles of curvature in biology.
#MedSky
Icecream: High-Fidelity Equivariant Cryo-Electron Tomography www.biorxiv.org/content/10.1101/2025.10.17.682746v1 #cryoEM
18.10.2025 08:50 — 👍 8 🔁 2 💬 0 📌 0
This comic is becoming increasingly realistic.
m.xkcd.com/3056/

𝗖𝘂𝗿𝗶𝗼𝘂𝘀 𝗵𝗼𝘄 𝘄𝗲 𝘂𝘀𝗲 𝗵𝗶𝗴𝗵-𝘁𝗲𝗰𝗵 𝘁𝗼 𝗮𝗱𝘃𝗮𝗻𝗰𝗲 𝗼𝘂𝗿 𝗿𝗲𝘀𝗲𝗮𝗿𝗰𝗵?
Then join us at our Open Day and get exciting sneak peaks behind the scenes of one of Europe’s largest life science campuses in Martinsried.
Find out more (in German): mpi.lineupr.com/tdot/
Time for a thread!🧵 How different is the molecular organization of thylakoids in “higher” plants🌱? To find out, we teamed up with @profmattjohnson.bsky.social to dive into spinach chloroplasts with #CryoET ❄️🔬. Curious? ..Read on!
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
gapTrick - Structural characterisation of protein-protein interactions using AlphaFold pubmed.ncbi.nlm.nih.gov/40985612/ #cryoEM
24.09.2025 22:31 — 👍 7 🔁 4 💬 0 📌 0hey bluesky 👋 visa hurdles mean I’m looking for opportunities outside the US. I’m a computational biologist (bacterial + phage genomics, postdoc in Koonin’s group @ NIH). I am interested in teaming up on funding apps. reach out if this resonates!
15.09.2025 17:26 — 👍 70 🔁 91 💬 1 📌 3Very impressive results and interesting approach. One can now wonder if the theoretical size limit still holds true. Has anyone tried to look at, e.g. lysozyme in cryoEM?
15.09.2025 17:16 — 👍 1 🔁 0 💬 1 📌 0This one is a bit of a departure from the usual and definitely a work in progress!
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x

In new study led by Xiaohui Ju, we define how mutations to Chikungunya virus envelope proteins affect entry in human vs mosquito cells.
Sheds light on functional constraints & enables us to make loss-of-tropism mutants, which could be of use for vaccines.
www.biorxiv.org/content/10.1...
Congrats Martin! Well deserved.
04.09.2025 14:34 — 👍 1 🔁 0 💬 0 📌 0
🌎👩🔬 For 15+ years biology has accumulated petabytes (million gigabytes) of🧬DNA sequencing data🧬 from the far reaches of our planet.🦠🍄🌵
Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...

First pre-print from the Lucas Lab led by Matthew Giammar and @joshdcryoem.bsky.social online today! We develop a new, extensible Python implementation of 2DTM and apply it to build pixel size optimization, constrained search and characterize molecular motions in situ www.biorxiv.org/content/10.1...
30.08.2025 00:49 — 👍 82 🔁 31 💬 8 📌 4
Sub-cellular chemical mapping in bacteria using correlated cryogenic electron and mass spectrometry imaging
Congrats Hannah Ochner and authors on this important paper! Strong collaboration with @kiranrpatil.bsky.social
www.biorxiv.org/cgi/content/...
@mrclmb.bsky.social @wellcometrust.bsky.social
Congratulations! Such an amazing and user friendly tool.
27.08.2025 22:09 — 👍 1 🔁 0 💬 0 📌 0
Better ML for cryo-ET starts with better benchmarks.
We built a phantom cryo-ET dataset (~500 tomograms) + hosted a Kaggle challenge.
The result: community models beat expert tools.
Read more in the @natmethods.nature.com article that just came out:
🔗 doi.org/10.1038/s415...
Researchers have developed a #DeepLearning system called BioEmu that rapidly generates diverse protein conformations, enabling fast, accurate insights into protein flexibility and function.
Learn more this week in Science: https://scim.ag/3JdQVD4
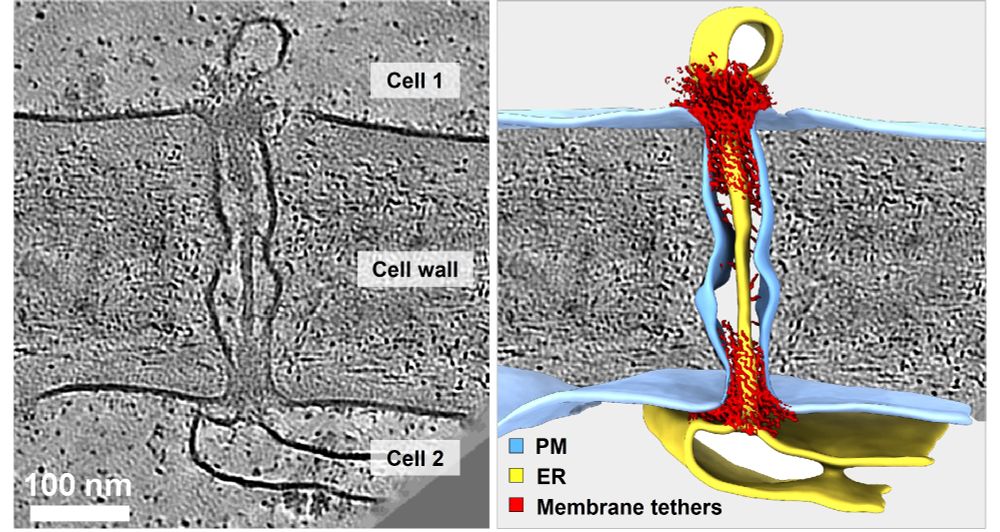
New preprint: In situ Architecture of #Plasmodesmata.
Using #cryoET, #AlphaFold & proteomics, we uncover the native organization of plant intercellular nanopores.
🔗 doi.org/10.1101/2025...
🧪🧵1/n
#teamtomo #PlantScience #Physcomitrium #Arabidopsis #cryoEM

Open Call for Expressions of Interest in Max Planck Directorships: Expressions of interest can be submitted until 31 October 2025.
Director at Max Planck - a unique position! The Open Call for Expressions of Interest in Max Planck Directorships is open now and can be submitted by the 31st of October 2025. ➡️ mpg.de/directors - Please share the Open Call among potential candidates.
01.08.2025 09:06 — 👍 162 🔁 196 💬 1 📌 15