Un viaggio all’origine dei sistemi nervosi
Il nuovo progetto GRNevo, finanziato con un grant FIS-3 Advanced da 1,9 milioni di euro, studierà come si formano i neuroni durante lo sviluppo in specie animali molto distanti tra loro: dal moscerino...
Thrilled to share that I’ve been awarded a FIS-3 Advanced Grant (ERC-inspired) to study the evolution of neurogenic GRNs.
Recruiting soon: 4 postdocs + 3 PhDs
Press release in Italian — to decolonise scientific language 😄
magazine.unibo.it/it/articoli/...
Email me if interested in joining the lab
13.01.2026 11:14 — 👍 61 🔁 15 💬 10 📌 1
Job offer 2026.pdf
🧬 We’re #Hiring a #Bioinformatician/ Computational Biologist!
#Single-cell genomics, #Epigenomics & #3Dgenome biology in #Zebrafish #DevBio 🐟
📍 @cabd-upo-csic.bsky.social, CABD, Seville (Spain)
🕒 Full-time, 3-year position (start March 2026)
📅 Apply by Feb 15, 2026
👉 drive.google.com/file/d/1M8rp...
12.01.2026 15:57 — 👍 17 🔁 22 💬 1 📌 2

Exciting news to start the new year! We’re thrilled to see this work finally out in @natecoevo.nature.com, the result of a major collaboration between my lab and the labs of Veronica Hinman and Nacho Maeso, began many years ago with the dear José Luis Gómez-Skarmeta.
www.nature.com/articles/s41...
07.01.2026 10:17 — 👍 40 🔁 16 💬 1 📌 3
(Cont. ; I'm very used to long posts from Mastodon): Thanks to @gacthel.bsky.social, Elena Emili, Anna Guixeras, Virginia Vanni, @drsalamander.bsky.social, @cireniasketches.bsky.social, Siebren Frölich, Simon Van Heeringen, Francesc Cebrià, @spiralcleavage.bsky.social & @jordisolana.bsky.social
27.11.2025 16:17 — 👍 2 🔁 0 💬 0 📌 0
We hope this work proves a valuable resource for the community (lots of classic functional experiments re-mapped to recent versions of the planarian genome), and helps paving the way into new hypotheses about which genes and regulatory elements mediate stem cell differentiation.
Once again thanks!
27.11.2025 16:07 — 👍 1 🔁 0 💬 1 📌 0
I learned so, so much in this project. Not only in terms of tools and analyses, but especially from the rest of the team, friends & peers at @jordisolana.bsky.social . Including critical thinking, how to coordinate a whole project -it's massive-, and how to communicate this research.
27.11.2025 16:07 — 👍 1 🔁 0 💬 1 📌 0
Just uploaded our new preprint to @ecoevorxiv.bsky.social
Multinucleate life really is everywhere...from deep sea forams to slime molds, intracellular parasites, and beyond. 🚀
We explore how scaling, ecology, & evolution intersect when many nuclei share a single cytoplasm.
doi.org/10.32942/X2M...
26.11.2025 15:06 — 👍 28 🔁 11 💬 0 📌 0
Happy to share Jialin's first publication. She did a great job exploring the transition to land in animals. Co-supervised by the great Jordi Paps and me and in collaboration with Davide Pisani and @phil-donoghue.bsky.social
13.11.2025 15:18 — 👍 64 🔁 34 💬 2 📌 3
Just gonna put this here for now
www.science.org/doi/10.1126/...
bsky.app/profile/jlst...
13.11.2025 19:23 — 👍 119 🔁 52 💬 14 📌 11

Picture of #LifelongDevSI Guest Editor Mansi Srivastava taken in an office with a whiteboard and sunny window. Mansi is wearing a white shirt and a flower pendant necklace with colourful petals
Mansi Srivastava is a Guest Editor for our Special Issue on Lifelong Development #LifelongDevSI. In this interview, Mansi talks about her research path, the links between regeneration and development and the exciting questions her lab is trying to answer.
journals.biologists.com/dev/article/...
16.10.2025 16:45 — 👍 18 🔁 4 💬 0 📌 0
congratulations @xgrau.bsky.social @arnausebe.bsky.social and the rest of the authors!
16.10.2025 12:02 — 👍 2 🔁 0 💬 1 📌 0

Latest from ours: www.cell.com/cell-reports...
This is two stories in one: a case study/cautionary tale on developing genetic tools in new organisms, and the first hint at a gene regulatory network for choanoflagellate multicellular development (which turn out to involve a Hippo/YAP/ECM loop!) A 🧵
05.10.2025 10:35 — 👍 242 🔁 101 💬 10 📌 10
Really grateful to see our work featured by @quantamagazine.bsky.social in this piece on the evolution of genome regulation. Huge thanks to @philipcball.bsky.social for such a beautifully written article.
08.10.2025 20:49 — 👍 36 🔁 18 💬 2 📌 0
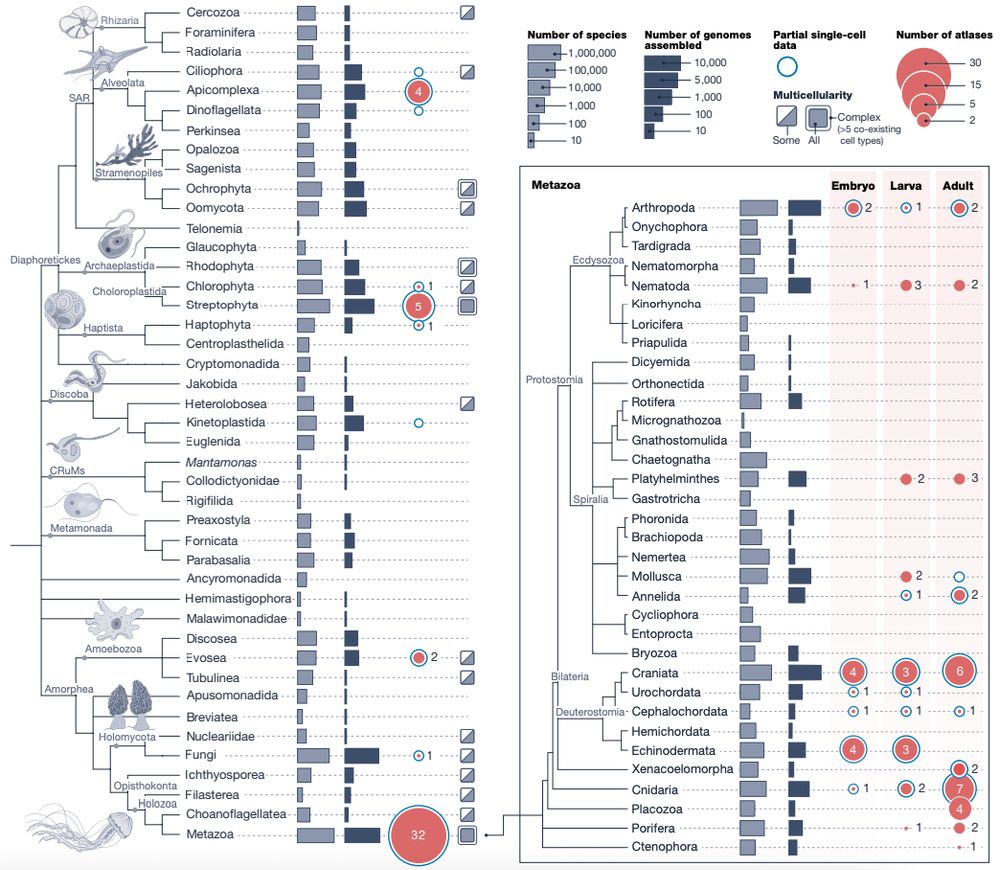
Happy to share the Biodiversity Cell Atlas white paper, out today in @nature.com. We look at the possibilities, challenges, and potential impacts of molecularly mapping cells across the tree of life.
www.nature.com/articles/s41...
24.09.2025 15:12 — 👍 226 🔁 106 💬 4 📌 10
Congratulations, amazing work! I remember when you explained to me the TF/motif strategy in the Sevilla meeting. I'm happy it worked well!
14.07.2025 09:44 — 👍 0 🔁 0 💬 0 📌 0
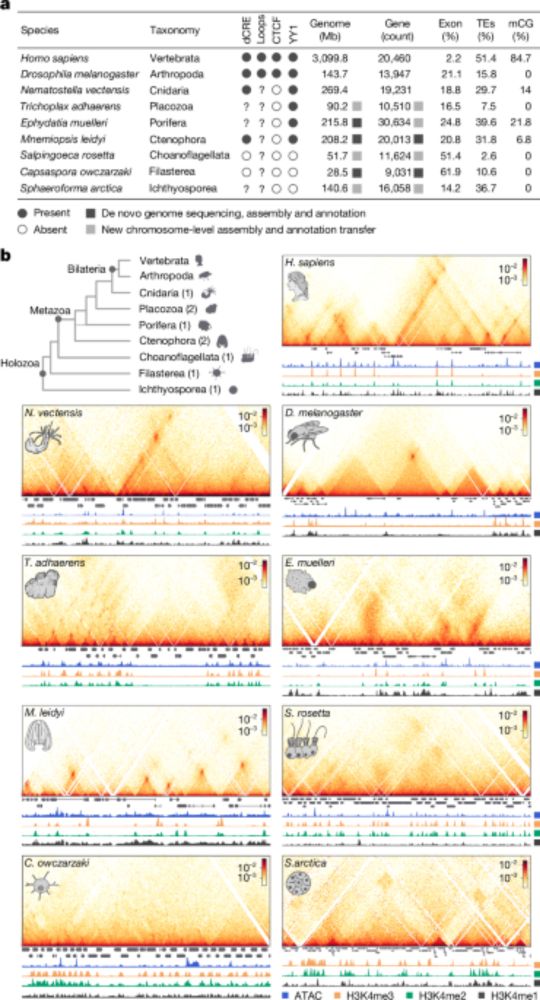
Decoding cnidarian cell type gene regulation
Animal cell types are defined by differential access to genomic information, a process orchestrated by the combinatorial activity of transcription factors that bind to cis -regulatory elements (CREs) to control gene expression. However, the regulatory logic and specific gene networks that define cell identities remain poorly resolved across the animal tree of life. As early-branching metazoans, cnidarians can offer insights into the early evolution of cell type-specific genome regulation. Here, we profiled chromatin accessibility in 60,000 cells from whole adults and gastrula-stage embryos of the sea anemone Nematostella vectensis. We identified 112,728 CREs and quantified their activity across cell types, revealing pervasive combinatorial enhancer usage and distinct promoter architectures. To decode the underlying regulatory grammar, we trained sequence-based models predicting CRE accessibility and used these models to infer ontogenetic relationships among cell types. By integrating sequence motifs, transcription factor expression, and CRE accessibility, we systematically reconstructed the gene regulatory networks that define cnidarian cell types. Our results reveal the regulatory complexity underlying cell differentiation in a morphologically simple animal and highlight conserved principles in animal gene regulation. This work provides a foundation for comparative regulatory genomics to understand the evolutionary emergence of animal cell type diversity. ### Competing Interest Statement The authors have declared no competing interest. European Research Council, https://ror.org/0472cxd90, ERC-StG 851647 Ministerio de Ciencia e Innovación, https://ror.org/05r0vyz12, PID2021-124757NB-I00, FPI Severo Ochoa PhD fellowship European Union, https://ror.org/019w4f821, Marie Skłodowska-Curie INTREPiD co-fund agreement 75442, Marie Skłodowska-Curie grant agreement 101031767
I am very happy to have posted my first bioRxiv preprint. A long time in the making - and still adding a few final touches to it - but we're excited to finally have it out there in the wild:
www.biorxiv.org/content/10.1...
Read below for a few highlights...
06.07.2025 18:14 — 👍 59 🔁 24 💬 1 📌 2
A single-cell ATACseq atlas of our favourite sea anemone, Nematostella vectensis! By @aelek.bsky.social @martaig.bsky.social
04.07.2025 10:27 — 👍 28 🔁 12 💬 1 📌 0

Congratulations to the new PhD at #CABD: Marta Moreno Oñate!
Dr. Moreno did an amazing job explaining the projects of her PhD work! Marta's thesis was done with @pablodeolavide.upo.es’s PhD program and was co-supervised by @martinfranke.bsky.social and Juan Tena!
01.07.2025 15:37 — 👍 3 🔁 1 💬 1 📌 1
I am very proud of you, Marta. You did outstanding work in the lab, and your defence was spotless. I am also very happy to now know an expert in gene compensation between paralogous genes!
02.07.2025 09:32 — 👍 2 🔁 2 💬 0 📌 0
Decoding cnidarian cell type gene regulation https://www.biorxiv.org/content/10.1101/2025.07.01.662323v1
03.07.2025 13:33 — 👍 4 🔁 3 💬 0 📌 1
And, if it turns out these long computation times are not due to lack of parallelisation, then there is something very strange going on that should be investigated. Functions like FindVariableFeatures, ScaleData, and others, are now reported to take several hours for 10K~30K cells datasets.
26.06.2025 15:45 — 👍 1 🔁 0 💬 0 📌 0
What is the current status of parallelisation support in Seurat? It was dropped last year with v5 and so far nothing has changed. It is not good that multiple people are reporting very slow runtimes (myself included).
github.com/satijalab/se...
github.com/satijalab/se...
github.com/satijalab/se...
26.06.2025 15:42 — 👍 1 🔁 0 💬 1 📌 0
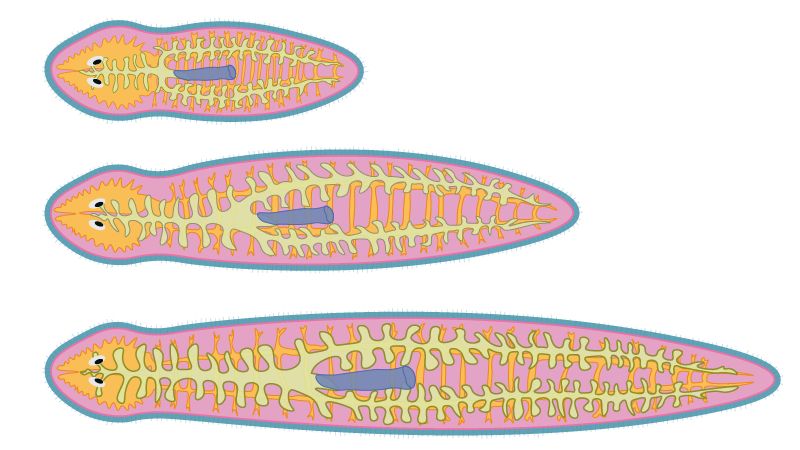
Proud to present the peer-reviewed version of our Cell Type Allometry paper, out today in Science Advances!
www.science.org/doi/10.1126/...
Are animals of different sizes made of the same cell types?
Here’s an update of the main points and revision items
(with memes!)
Thread 👇🧵
07.05.2025 19:01 — 👍 95 🔁 35 💬 6 📌 3

Lungfish xkcd.com/3064
17.03.2025 16:53 — 👍 11936 🔁 1308 💬 96 📌 87

Y'all so excited to build this world lol
17.03.2025 22:57 — 👍 34 🔁 7 💬 2 📌 0
Evolutionary Biologist at UCSC | Assistant professor | Interested in genes, genomes, and #evolution of complex traits 🧬 | EvoDevo Regeneration
https://macias.sites.ucsc.edu/
📸 IG: @cnidolab_ucsc
Group leader at CABD (Seville). Previous at Max Planck Institute for Molecular Genetics (Berlin). Interested in evolutionary genomics of moles and in any other living creature!
Email: fmarrea@upo.es
Web: https://fmreallab.github.io/website/
Mathematics and biology enthusiast
Evolutionary Biologist working on annelida developmental biology at Universidad Autonoma de Madrid @Biología UAM• #annelids #evolution #reproduction #regeneration
Postdoctoral researcher in the Sebe-Pedros and Marti-Renom Labs at CRG. Transposable elements enthusiast, passionate about piRNAs, 3D genomes, and Star Trek 🖖
Evolutionary biologist exploring the molecular mechanisms behind life's complexity. Currently @International University of Catalonia (UIC), Barcelona.
Theoretical Physicist.
(She/her/hers)
From Eastern KY.
Aging and cancer stem cell heterogeneity - ICREA research professor - Quantitative Stem Cell Dynamics lab
at IRB Barcelona 🇦🇷🇪🇺🇺🇸 fraticellilab.com
The Epigenetic Face of Cancer Metabolism Lab
We study NUCLEAR METABOLISM at CRG, Barcelona
https://sdelcilab.crg.eu/
Asst Professor, School of Biomedical Engineering, UBC; stem cell engineer and synthetic biologist with a passion for science outreach and communication.
https://shakiba.bme.ubc.ca/
I am a cell and developmental biologist at the Living Systems Institute (LSI) in Exeter. We work on spreading Wnt signals and characterising cytonemes in zebrafish embryos, human neurons, and cancer using high-res imaging and mathematical modelling.
Agencia Estatal de Investigación de España. Fomentamos la investigación científica y técnica mediante la asignación de los recursos públicos.
Paleontologist. Developmental Biologist. Anatomist. Polar wanderer. Telling people that they are fish since 2008.
2025 Schmidt Science Fellow
CDB UCL London and Gurdon Institute Cambridge
Wilson, Simons and Norden labs
Developmental, Systems and Synthetic Biology
Prev: Briscoe, Crick Institute; Zernicka-Goetz and Thomson, Caltech; Banerjee and Goehring, UCL
🧬 3D genomics in evo-devo 🪰
🧫 3D'omics modeling @3domics.bsky.social
Bioinformatician @mamartirenom.bsky.social lab, CNAG, Barcelona
PhD student on leave in the Prof. Gelfand lab at Skoltech, Moscow
PhD student in the Sebé-Pedrós lab at CRG.
Postdoc at the Department of Natural History, @universitetsmuseet.bsky.social, @unibergen.bsky.social | Interested in diversity, ecology and life cycles of Cnidaria and Ctenophora 🪼🪸|