So happy to announce our new preprint, “A geothermal amoeba sets a new upper temperature limit for eukaryotes.” We cultured a novel amoeba from Lassen Volcanic NP (CA, USA) that divides at 63°C (145°F) 🔥 - a new record for euk growth!
#protistsonsky 🧵
25.11.2025 20:41 — 👍 478 🔁 164 💬 16 📌 29
How do cells adapt morphology to function? In a 🔥 preprint by @zjmaggiexu.bsky.social , with @dudinlab.bsky.social and @amyweeks.bsky.social , we identify a self-organizing single-cell morphology circuit that optimizes the feeding trap structure of the suctorian P. collini. 🧵 tinyurl.com/4k8nv926
18.11.2025 16:15 — 👍 130 🔁 55 💬 4 📌 11

🌍Open call: Junior Group Leader positions!
Join a world-class biomedical research institute at the heart of the Vienna BioCenter, where curiosity drives discovery.
Lead your own lab, pursue bold ideas, and shape the future of science at the IMP: www.imp.ac.at/career/open-...
10.11.2025 13:26 — 👍 95 🔁 86 💬 1 📌 7
How is epigenetic information inherited? We found that CDCA7 proteins are critical players in the inheritance of DNA methylation at CG sites in plants, and this is true both in the lab and in the wild. How does this work? 🧵👇
07.11.2025 17:01 — 👍 43 🔁 15 💬 2 📌 1
🚨Our collaboration with @centriolelab.bsky.social & @gautamdey.bsky.social is out today in @cp-cell.bsky.social
We show that #Expansion #Microscopy is a broad-spectrum modality for Euks, enabling 3D phenotypic maps rooted to phylogeny.
#ProtistsOnSky #SciComm #SciSky
www.cell.com/cell/fulltex...
31.10.2025 14:41 — 👍 215 🔁 99 💬 15 📌 11

Lab’s first paper is out!! We show the first structures of #Asgard #chromatin by #cryo-EM 🧬❄️
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
28.10.2025 15:07 — 👍 329 🔁 115 💬 6 📌 8
New preprint! We unexpectedly discovered that some Caenorhabditis species delete parts of their somatic genome early in development, which fragments their chromosomes and eliminates key germline genes. Multiple lines of evidence suggest this bizarre process was present in the ancestors of C. elegans
28.10.2025 12:11 — 👍 49 🔁 17 💬 0 📌 0
The CRG PhD call is now open. Exciting opportunities across diverse topics and within a world-class scientific environment.
Our group is offering one PhD position to study chromatin evolution.
Consider applying or share with anyone who might be interested!
www.crg.eu/en/content/t...
23.10.2025 11:54 — 👍 18 🔁 15 💬 0 📌 0

Picture of an Oculina patagonica colony. Some of the individual polyps in the colony have a brown-yellow colour, indicating that they harbour symbiotic algae. Others are completely white and lack algae.

Close-up picture of symbiotic Oculina polyps.
Hot off the press! Our latest work on the evolution of facultative symbiosis in stony corals, focusing on a remarkable Mediterranean species: Oculina patagonica.
🪸 🌊
#evobio #corals #coralbiology
www.nature.com/articles/s41...
15.10.2025 20:29 — 👍 46 🔁 11 💬 2 📌 0

Opportunity for a Master’s/Bachelor’s student:
- Join us for up to 5 months 🗓️
- Build computational/mathematical models 💻
- Learn about genotype-phenotype maps and evolution 🧬
- Work closely with PhD student Manuela Giraud - full info here:
www.crg.eu/en/content/t...
07.10.2025 14:38 — 👍 8 🔁 7 💬 0 📌 0
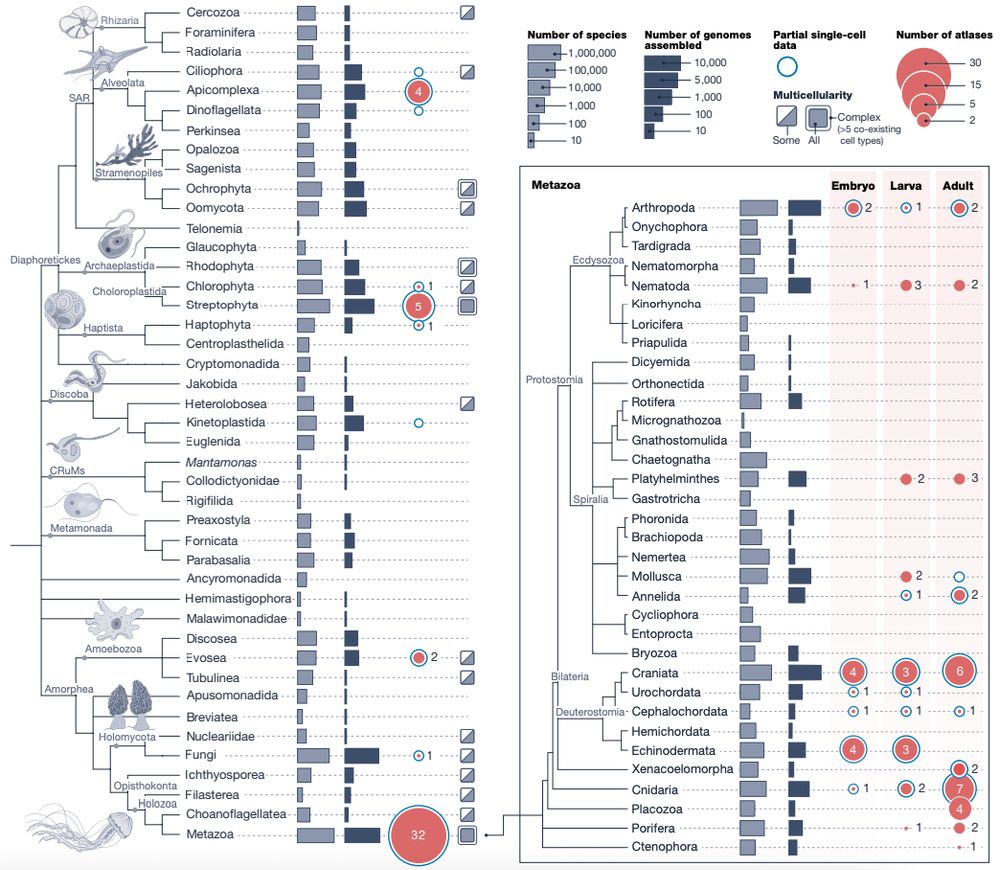
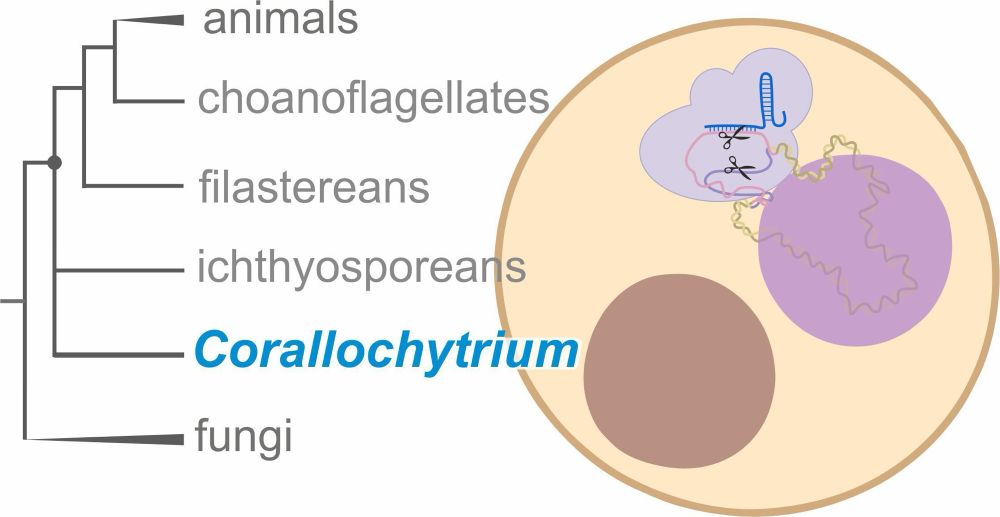
Happy to share the Biodiversity Cell Atlas white paper, out today in @nature.com. We look at the possibilities, challenges, and potential impacts of molecularly mapping cells across the tree of life.
www.nature.com/articles/s41...
24.09.2025 15:12 — 👍 225 🔁 106 💬 4 📌 10
Brown algae up to some interesting strategies regarding gene repression, losing all "traditional" pathways while expanding the DOT1 (H3K79me writer) protein family! I hope to learn more at the EMBO workshop next month in Barcelona!
22.09.2025 13:33 — 👍 6 🔁 1 💬 0 📌 0

Activity of most genes is controlled by multiple enhancers, but is there activation coordinated? We leveraged Nanopore to identify a specific set of elements that are simultaneously accessible on the same DNA molecules and are coordinated in their activation. www.biorxiv.org/content/10.1...
18.08.2025 12:23 — 👍 97 🔁 39 💬 2 📌 2
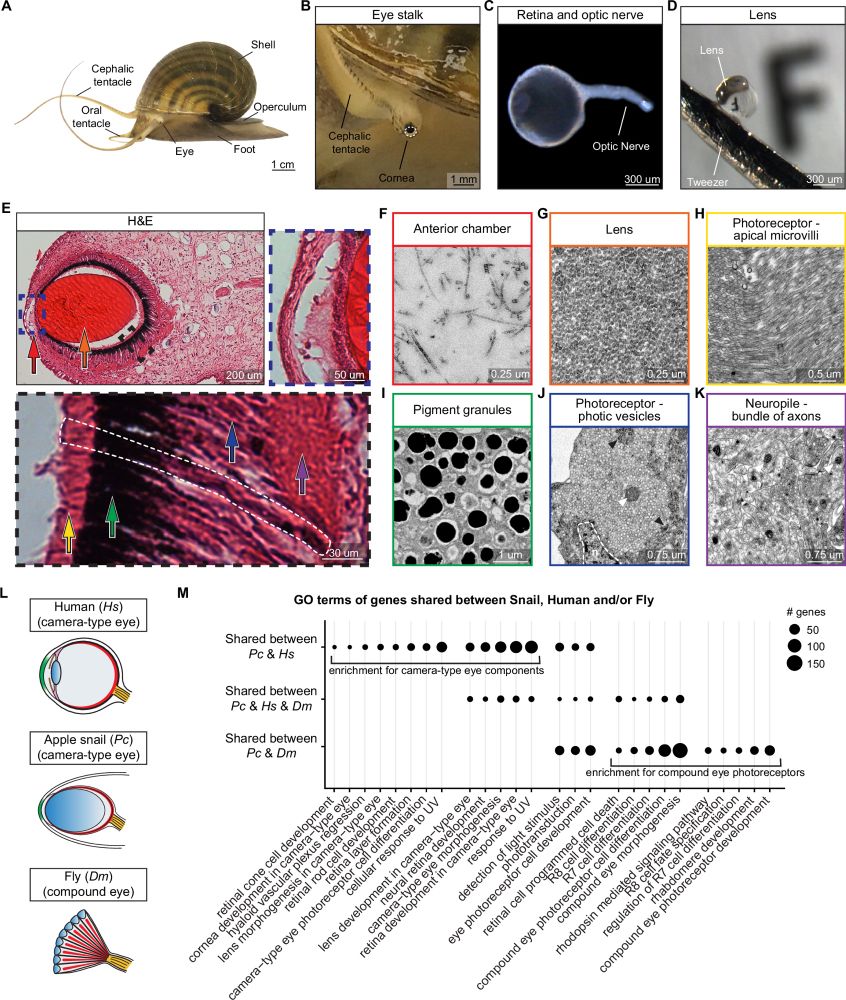
A genetically tractable non-vertebrate system to study complete camera-type eye regeneration
Nature Communications - Accorsi et al. show that the apple snail Pomacea canaliculata has eyes similar to humans and can fully regenerate them. They then developed genetic tools to establish these...
Evolution’s eye game is wild, but mollusks take it to another level
CRISPR in apple snails gives us a new model to dissect how nature rebuilds complex organs like the camera-type eyes we humans possess
It turns out Evolution doesn’t just innovate, it rewinds, remixes, & regenerates
rdcu.be/ezw0t
06.08.2025 11:21 — 👍 117 🔁 43 💬 1 📌 2

Academic poaching: when other PIs steal your trainees
There are many toxic behaviors in academia, but one we rarely talk about is #AcademicPoaching. Have you ever been a victim?
I just published: Academic poaching: when other PIs steal your trainees
There are many toxic behaviors in academia, but one we rarely talk about is #AcademicPoaching. Have you ever been a victim?
#AcademicChatter #AcademicSky #phdchat
medium.com/p/academic-p...
28.07.2025 13:00 — 👍 27 🔁 11 💬 3 📌 2
And another opportunity! Any postdocs looking forward to start your lab on a tenure track? Supportive environment, great core facilities and an outstanding PhD program.
23.07.2025 13:42 — 👍 9 🔁 9 💬 0 📌 0
We are looking for new colleagues to come join us in Galway as group leaders (Junior and Senior). The Centre for Chromosome Biology is a great place and it is a good time to join. Please reach out if you want to chat about the opportunity!
www.nature.com/naturecareer...
16.07.2025 08:56 — 👍 67 🔁 56 💬 2 📌 2
Pls. share widely
Calling all transposon fans & lovers of genetic innovation
MOBILE GENOME welcomes you in Heidelberg, Nov. 4–7 2025
→ Vibrant & friendly community
→ Cutting-edge talks from mechanisms to physiology
→ Plenty of surprises (TEs never stop innovating)
submit abstract by July 29
16.07.2025 08:43 — 👍 61 🔁 48 💬 1 📌 1

We are looking for a student to continue our work on chromatin evolution:
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
The project with @seanamontgomery.bsky.social will focus on chromatin state readers across eukaryotes.
More info: recruitment.crg.eu/content/jobs...
15.07.2025 11:30 — 👍 28 🔁 38 💬 0 📌 1

Sorry, the ad image didn't look so good in dark mode, here it is again!
15.07.2025 07:16 — 👍 0 🔁 0 💬 0 📌 0
Incoming Group Leader @ MPI-CBG
Postdoc @ Schier lab
PhD @ Ameres lab
Fascinated about post-transcriptional gene regulation & RNA biology in early development.
A.d.i.d.a.s. All day I dream about science. Group Leader at IMBA. Genomics & evolution. Peruano.
Scientist at IMP in Vienna. Excited about gene expression regulation and its encoding in our genomes - enhancers, transcription factors, co-factors, silencers, AI.
Group leader at IMBA Vienna | Excited about #devbio #synbio #stemcells #patterning #morphogenesis #metabolicsignalling #energetics | Investigating how metabolism shapes development
https://www.oeaw.ac.at/imba/groups/kristina-stapornwongkul
Interested in the organisation of chromatin in bacteria and archaea, its interplay with transcription and evolution. Methods: single-molecule and ensemble biochemistry, genetics, molecular microbiology, 3D genomics, (computational) structural biology.
Group Leader at EMBL spying on the social life of marine microbes and pushing for more inclusive sciences | Tara Oceans | Plankton Manifesto
https://www.embl.org/groups/vincent/
https://planktonmanifesto.org/
Scientist & Communicator. Fascinated by cells across scales - 🌎, 🔬and ⏳. PI @GIMM, EMBO member. Former Director @IGC, PD @Cambridge University, PhD @UCL/IGC. My motto: "Science from all for all". Mom of 2 wonderful girls. Views are my own.
🇩🇿 🇫🇷 scientist @Inserm - https://iab-grenoble.fr/
👀 Host-Parasite Coevolution: The Toxoplasma Paradigm
ParaFrap deputy director - https://www.labex-parafrap.fr/en/
EMBO member - https://www.embo.org/
🌍 pan-African idealist
#epigenetics #parasites
We use T. brucei to study the role of chromatin and genome architecture in antigenic variation.
https://www.vetmed.lmu.de/molpara/en/
Postdoc in the Pauli Group @IMP in Vienna. Interested in gene expression regulation during early embryonic development in 🐟
Sequencing microscopic life one cell at a time @earlhaminst.bsky.social. Documenting microscopic life one pond at a time @pondlife_pondlife.
Group leader at @ibe-barcelona.bsky.social (@csic.es - @upf.edu) & Adjunct at @miamirosenstiel.bsky.social. Microbial Ecology & Evolution 🔬🌍🧫🧬 🪸 @pr2-database.bsky.social Coordinator #corals #symbiosis #protists
Biophysicist | genotype-phenotype maps, fitness landscapes and evolution | Independent Fellow leading a group at @barcelonacollaboratorium.com @crg.eu
Interested in evolutionary microbiology, symbiosis, organelles and protists. Currently an MSCA postdoc at HHU, Düsseldorf.
ꙩ ꙫ ө ꚛ ꙮ ༗ :: complex multicellularity in brown algae :: doctoral researcher :: max planck institute for biology :: tübingen
https://lotharukpongjs.github.io/
Postdoc @ Garvan Institute | Epigenetics, development, evolution
Professor for Genetics interested in chromatin structure, histone variants and histone modifications and their roles in gene regulation and disease development.
Assistant Prof and Group Leader at Max Perutz Labs, University of Vienna
Doctoral researcher @mpi-bio-fml.bsky.social and curious human being
De-coding brown algae 🍙