Zero-Shot De Novo Peptide Sequencing with Open Post-Translational Modification Discovery www.researchsquare.c...
---
#proteomics #prot-preprint
@jeroen.vangoey.be
Staff Research Engineer in BioAI at InstaDeep (part of @biontech.bsky.social)- machine learning for personalized cancer vaccines, de novo peptide sequencing and signal peptides. From Belgium 🇧🇪 currently living in Cape Town 🇿🇦 #bioML #TeamMassSpec

Zero-Shot De Novo Peptide Sequencing with Open Post-Translational Modification Discovery www.researchsquare.c...
---
#proteomics #prot-preprint
Xiang Zhang, Jiaqi Wei, Zijie Qiu, Sheng Xu, Nanqing Dong, Zhiqiang Gao, Siqi Sun: Curriculum Learning for Biological Sequence Prediction: The Case of De Novo Peptide Sequencing https://arxiv.org/abs/2506.13485 https://arxiv.org/pdf/2506.13485 https://arxiv.org/html/2506.13485
17.06.2025 09:25 — 👍 2 🔁 2 💬 0 📌 0
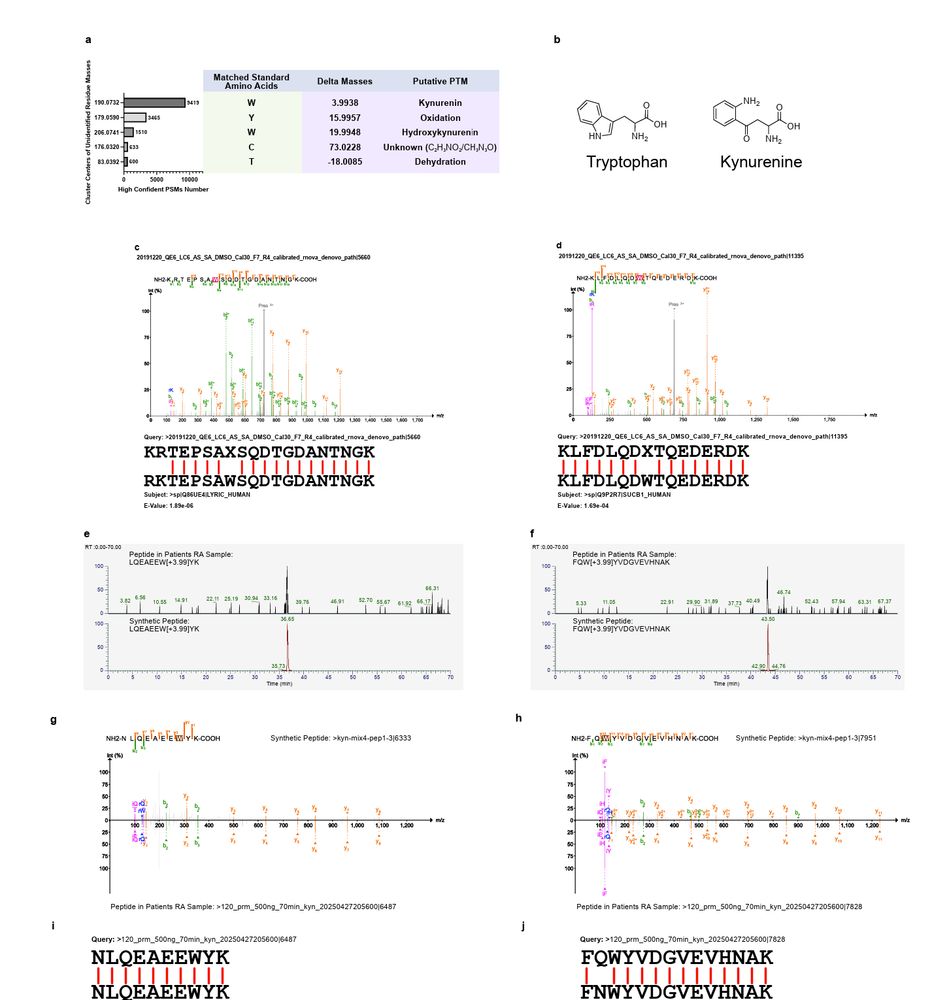
Figure 2
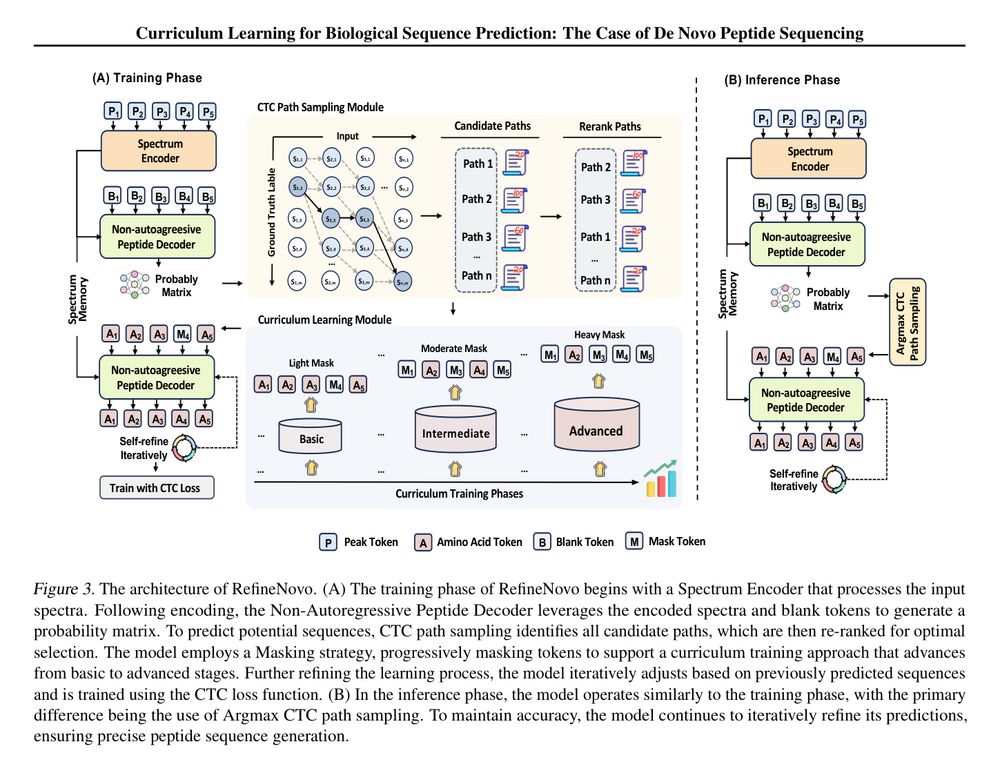
Figure 3
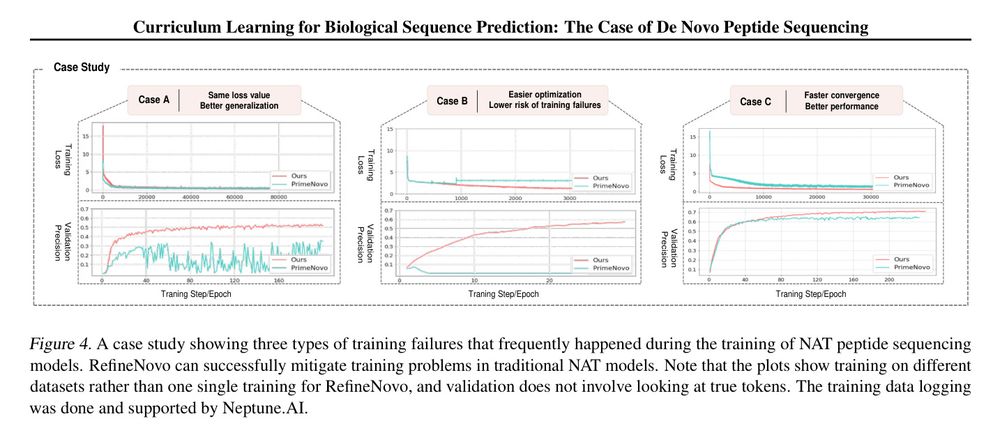
Figure 4
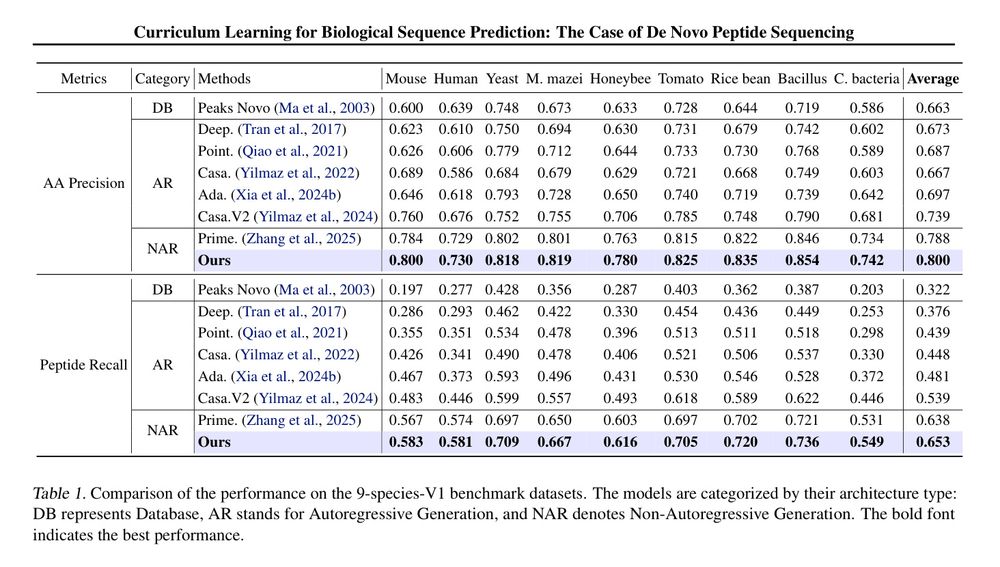
Table 1
Curriculum Learning for Biological Sequence Prediction: The Case of De Novo Peptide Sequencing [new]
Curriculum Learning enhances non-autoregressive peptide sequencing using structured training, adjusting difficulty for refined predictions.

🚨 New preprint! 🚨
Presenting diaPASEF immunopeptidomics for bacterial epitope discovery.
💥 Showcasing DIA-NN immunopeptidomics using proteome-wide predicted HLA class I libraries!
🧬 Read more: biorxiv.org/cgi/content/...
#Immunopeptidomics #MassSpec #DIA #HLA

(BioRxiv All) Data-independent immunopeptidomics discovery of low-abundant bacterial epitopes: Mass spectrometry-based immunopeptidomics is a powerful approach to uncover peptides presented by human leukocyte antigen (HLA) molecules that can guide vaccine design and… #BioRxiv #MassSpecRSS
16.05.2025 06:58 — 👍 7 🔁 1 💬 0 📌 0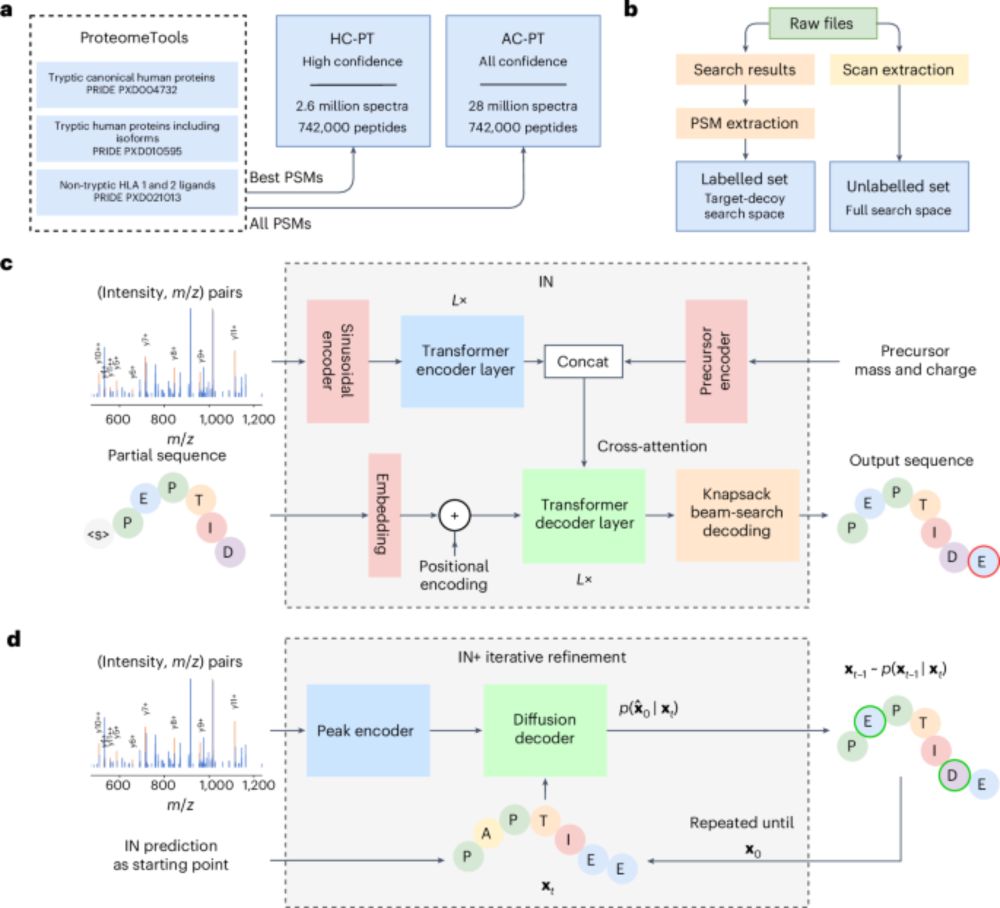
'We uncover novel biological findings across eight different datasets, including the identification of proteins in HeLa cells undetected by database search, the expansion of the immunopeptidomics dataset by 175% more peptides and the characterization of novel proteolytic cleavages.'
05.04.2025 09:59 — 👍 3 🔁 2 💬 0 📌 0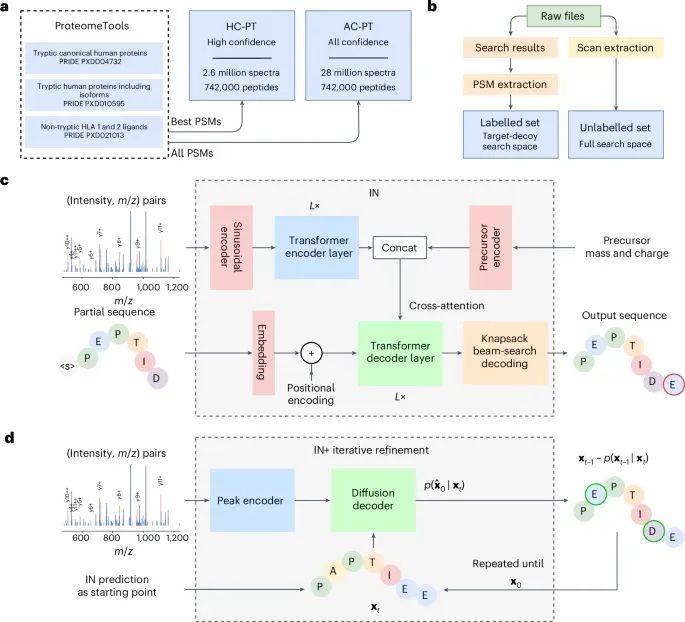
InstaNovo enables diffusion-powered de novo peptide sequencing in large-scale proteomics experiments - Nature Machine Intelligence #DL #AI #ML #DeepLearning #ArtificialIntelligence #MachineLearning
www.nature.com/articles/s42...
Open-Source and FAIR Research Software for Proteomics pubs.acs.org/doi/10....
---
#proteomics #prot-paper
A review from the Journal of #Proteome #Research | Open-Source and #FAIR Research Software for #Proteomics | #Bioinformatics #OpenScience #OpenSource 🖥️ 🧪 🔓
⬇️
pubs.acs.org/doi/10.1021/...
Fantastic review with an unusual history, growing out of a passionate blog post by @willfondrie.com (willfondrie.com/2024/10/the-...), resulting from a storm (in our teacup) on X during @hupo-org.bsky.social 2024. Great teamwork, authors! pubs.acs.org/doi/10.1021/...
24.04.2025 18:24 — 👍 16 🔁 11 💬 0 📌 1
Use a template like github.com/greenelab/la... and publish to GitHub Pages
20.04.2025 15:47 — 👍 1 🔁 0 💬 0 📌 0
The AI revolution comes to protein sequencing www.science.org/cont...
---
#proteomics #prot-article

Are you looking for something cool to read this weekend? 😎 What about using AI to improve protein sequencing? It's an exciting one! 🧪🧬🖥️ plentyofroom.beehiiv.com/p/ai-protein...
18.04.2025 16:12 — 👍 9 🔁 2 💬 0 📌 1
New issue today! 🎉 I covered a great article out of @dtu.dk and @instadeep.bsky.social: can AI accelerate peptide discovery? 😎🧪🖥️🧬 The answer is yes, thanks to InstaNovo! Read everything and subscribe at 👉 plentyofroom.beehiiv.com/p/ai-protein... #AI #machinelearning #biotech #proteinscience
17.04.2025 15:41 — 👍 6 🔁 1 💬 0 📌 1
2/
What’s new in InstaNovo v1.1?
🧬 +13.5% recall
🎯 +42.6% more exact PSMs
🔍 145% more peptides & 35% more proteins @ 5% FDR
🧪 Support for 7 PTMs: phosphorylation, deamidation, carbamylation, ammonia loss, oxidation, acetylation, carbamidomethylation
Paper: bit.ly/3XBeJFh

1/
🚀 InstaNovo just got a major upgrade.
Our Nature Machine Intelligence paper with @instadeepai presents v0.1—but while the paper was under review, we kept building.
Now releasing InstaNovo v1.1, with major gains.
Blog: bit.ly/4lircYB
🧵

New AI Tool Unlocks Hidden Proteins in Sequencing Breakthrough Artificial intelligence (AI) has b...
https://www.hpcwire.com/2025/04/10/new-ai-tool-unlocks-hidden-proteins-in-sequencing-breakthrough/
#Short #Takes #AlphaFold #Casanovo #folding #InstaNovo […]
[Original post on hpcwire.com]

The AI models, called InstaNovo and InstaNovo+, are a step toward “the holy grail” of protein research: to unravel the genetic identity of previously unstudied proteins en masse
12.04.2025 13:10 — 👍 6 🔁 3 💬 0 📌 0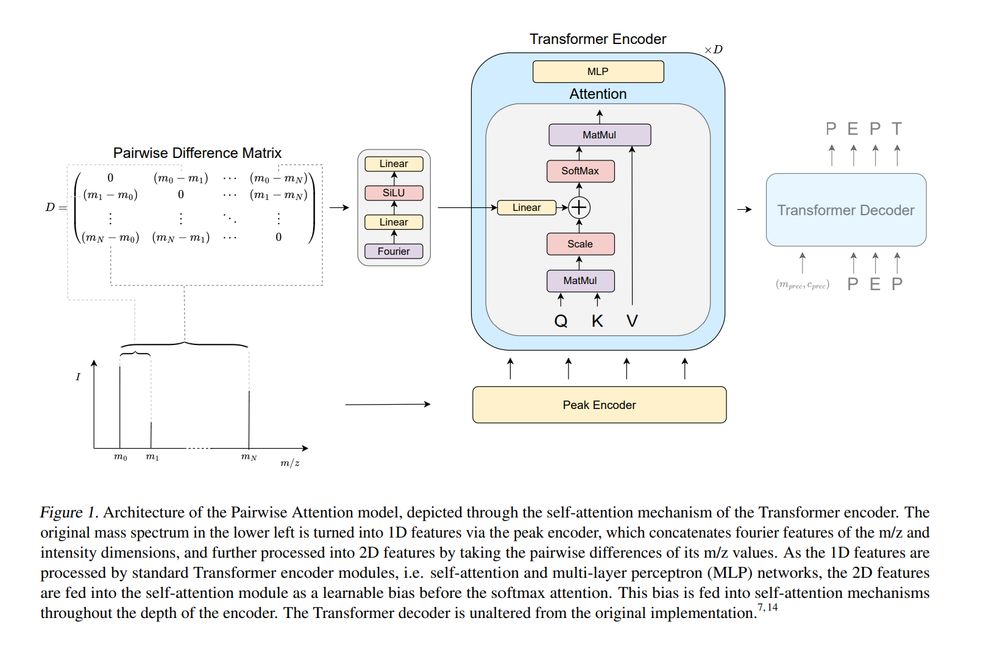
Pairwise Attention: Leveraging Mass Differences to Enhance De Novo Sequencing of Mass Spectra www.biorxiv.org/cont...
---
#proteomics #prot-preprint
OpenAI, Perplexity and German drugmaker BioNTech and its London-based AI subsidiary InstaDeep have recently launched their own AI research tools, while Google DeepMind’s AlphaFold has shown how the fast-developing technology can accelerate scientific research.
20.02.2025 00:22 — 👍 2 🔁 1 💬 0 📌 0Seconding putting the code and requesting a DOI. You can also add a CITATION.cff file to your repository.
citation-file-format.github.io
Or put the code on zenodo.org, then you also get a DOI.
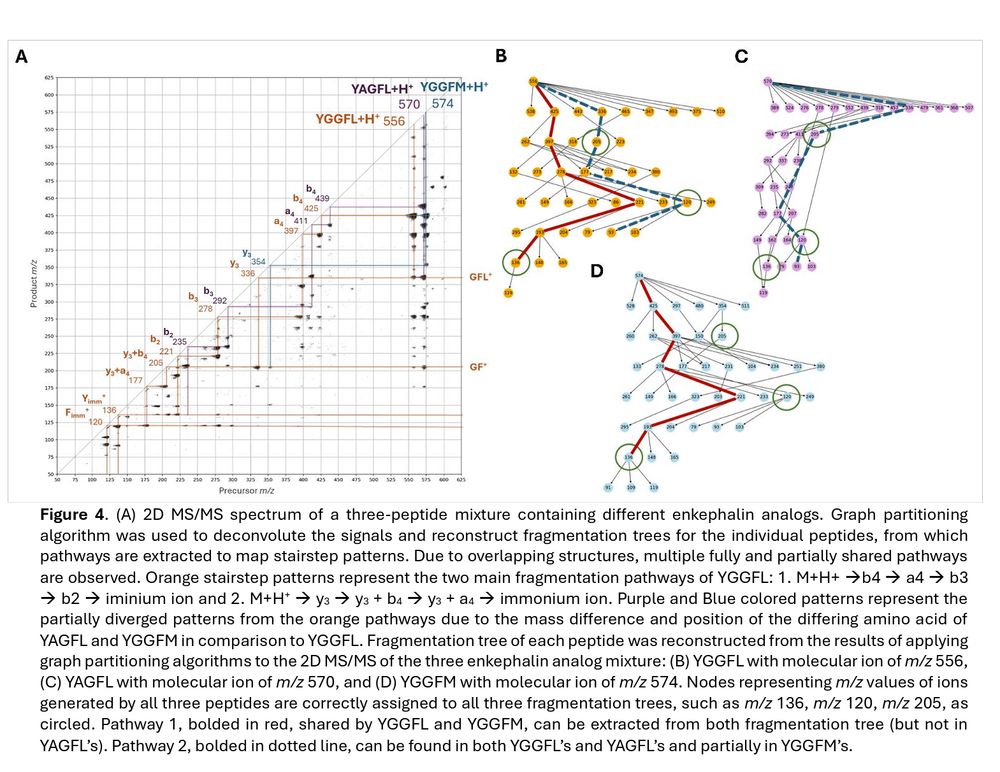
Framework for De novo Sequencing of Peptide Mixtures via Network Analysis and Two-Dimensional Tandem Mass Spectrometry chemrxiv.org/engage/...
---
#proteomics #prot-preprint
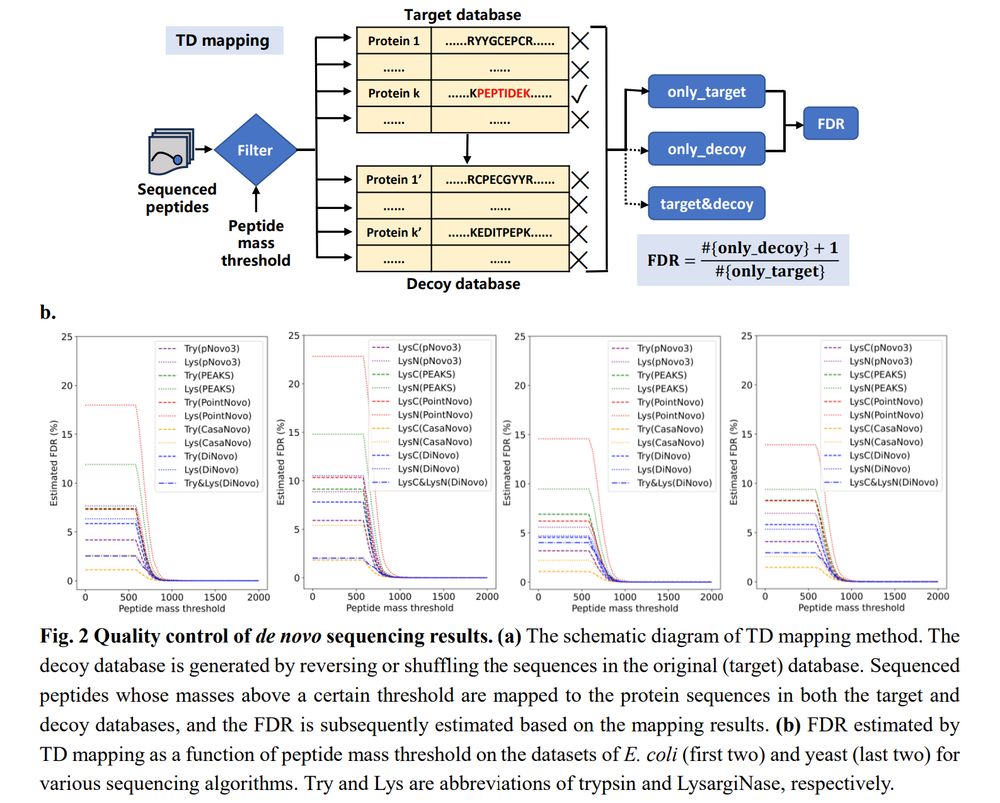
DiNovo: high-coverage, high-confidence de novo peptide sequencing using mirror proteases and deep learning www.biorxiv.org/cont...
---
#proteomics #prot-preprint
DiNovo: high-coverage, high-confidence de novo peptide sequencing using mirror proteases and deep learning https://www.biorxiv.org/content/10.1101/2025.03.20.643920v1
25.03.2025 15:47 — 👍 4 🔁 2 💬 0 📌 0
Pairwise Attention: Leveraging Mass Differences to Enhance De Novo Sequencing of Mass Spectra
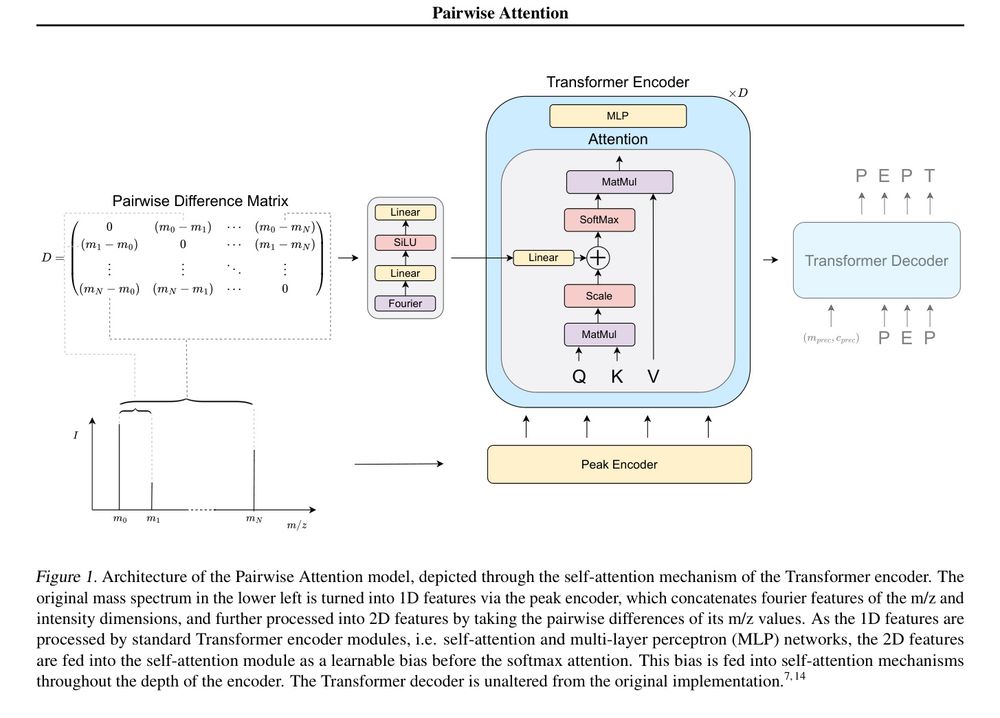
Figure 1
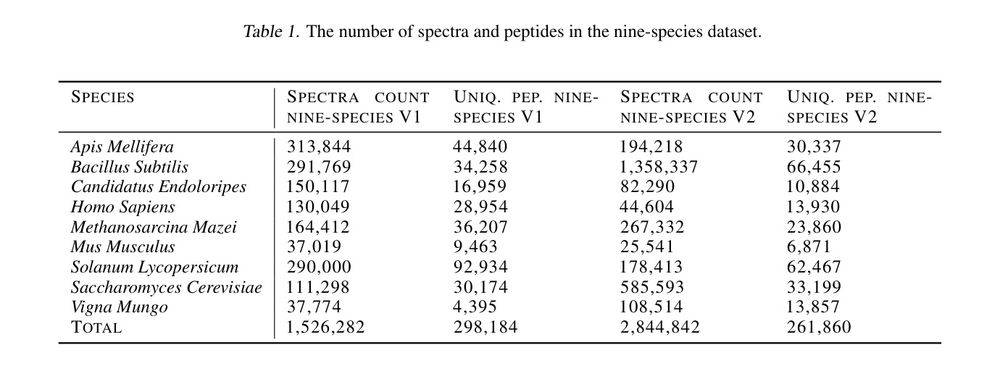
Table 1
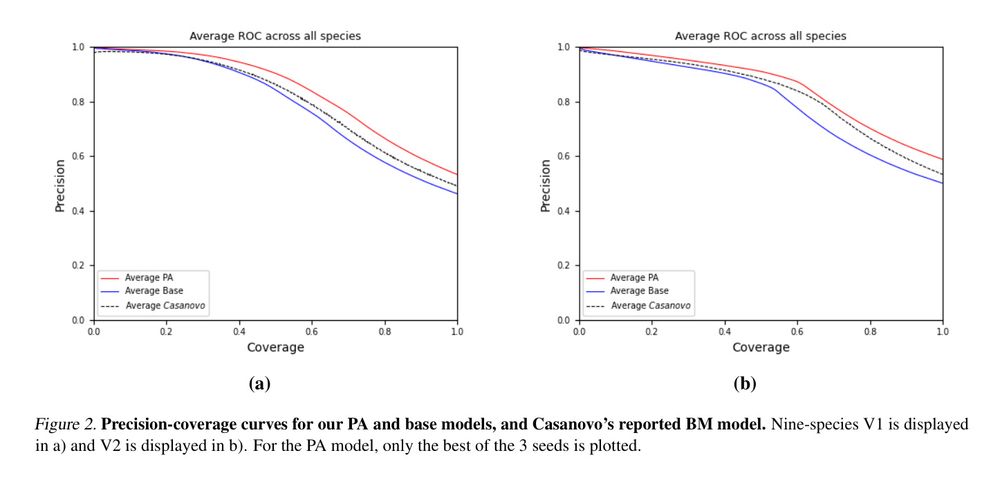
Figure 2
Pairwise Attention: Leveraging Mass Differences to Enhance De Novo Sequencing of Mass Spectra [new]
Use mass differences to improve transformer networks for peptide sequencing by biasing the attention mechanism.
Google colab has free GPUs (T4). Instanovo now has a Hugging Face space for online predictions! CPUs still work for Casa & Insta (just slower🐢)
03.04.2025 07:44 — 👍 3 🔁 1 💬 1 📌 0
#AI tools InstaNovo and InstaNovo+ decode hidden proteins missed by traditional methods, aiding #cancer research & understanding diseases.
These tools significantly outperform conventional protein detection, offering potential for new therapies.
Source: shorturl.at/6nGkc
#Health #Research
'‘InstaNovo … directly predicts the sequence from the spectrum, eliminating the need for database lookups (...) This is possible because our models have learned the underlying patterns of the sequences we are measuring and can translate a spectrum directly into the corresponding peptide sequences.’'
05.04.2025 09:57 — 👍 4 🔁 7 💬 1 📌 0
"Over the past 4 years, researchers have unveiled more than two dozen protein sequencing AIs. “It seems clear that this is where the field is going to go,” says William Noble, a proteomics AI developer at the University of Washington."
04.04.2025 08:03 — 👍 4 🔁 1 💬 0 📌 0
Considering how old the mass spec is in this picture , they likely need all the ai they can get to get good data out of that thing !
www.science.org/content/arti...