The #mattertolife fall days are a wonderful opportunity to connect to this vibrant community. This time at the @mpip-mainz.mpg.de with talks by the local hosts @graeterlab.bsky.social, @landfesterdept.bsky.social and @weilgroup.bsky.social. Thanks @mattertolife.bsky.social for bringing us here 👍😀🙏
22.09.2025 16:14 — 👍 12 🔁 2 💬 0 📌 0

Lecture hall with a large group of people watching a presentation
Our #mattertolife Fall Days have just started 👏🏻 Thank you @mpip-mainz.mpg.de for hosting us!
@graeterlab.bsky.social @landfesterdept.bsky.social @weilgroup.bsky.social
22.09.2025 07:12 — 👍 9 🔁 2 💬 0 📌 0
Flabby, flexible, and full of potential: Researchers at HITS and @mpip-mainz.mpg.de have developed a #machinelearning model that designs flexible proteins – even with rare patterns. A leap toward next-gen enzymes, therapeutics & sustainable biotech! 🌱💊🔬
👉 Read more: www.h-its.org/2025/07/23/f...
23.07.2025 12:47 — 👍 20 🔁 5 💬 0 📌 0
Want your de novo designed protein to be wobbly? 🍮 Seva, Leif & team have made it possible - you find them at ICML! 🤩 @mpip-mainz.mpg.de @hitsters.bsky.social
17.07.2025 18:29 — 👍 12 🔁 2 💬 0 📌 0
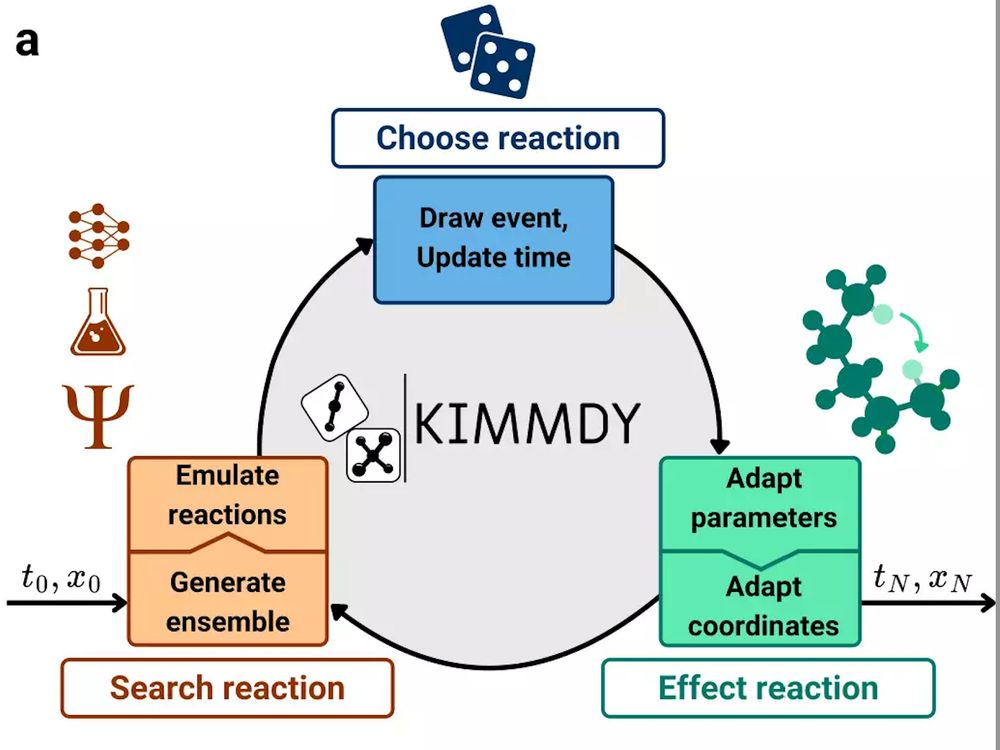
You want to make your simulations reactive, at little cost? Try KIMMDY. Don't hesitate to reach out if you want to include your favorite chemistry. Super happy to see this online, kudos to the team!
www.biorxiv.org/content/10.1...
07.07.2025 07:00 — 👍 7 🔁 1 💬 0 📌 0
Congrats, Arne!
19.05.2025 10:09 — 👍 0 🔁 0 💬 1 📌 0
Join us at the MPIP in Mainz - a few days left to apply! You feel this matches your research agenda but would be a step too early in your career? Do apply!
09.05.2025 05:30 — 👍 5 🔁 1 💬 0 📌 0
@rebecca-wade.bsky.social @graeterlab.bsky.social @janstuehmer.bsky.social @astroinformatics.bsky.social @leif-seute.bsky.social
07.05.2025 13:00 — 👍 4 🔁 4 💬 0 📌 1
Fantastic news, congratulations, Pascal!
28.04.2025 09:26 — 👍 0 🔁 0 💬 0 📌 0

Registration Deadline Extended!
The SIMPLAIX Workshop 2025 is just around the corner, and there's still time to join us!
📅 New deadline: 13 April 2025
📍 Studio Villa Bosch, Heidelberg
🔗 Register now: simplaix-workshop2025.h-its.org
Spots are filling fast — secure yours today! #SIMPLAIX25
07.04.2025 12:49 — 👍 7 🔁 5 💬 0 📌 0
Kai's work among 'Most popular articles 2024' in Chem Sci - many thanks to the readers :-)
tinyurl.com/4yb78j5e
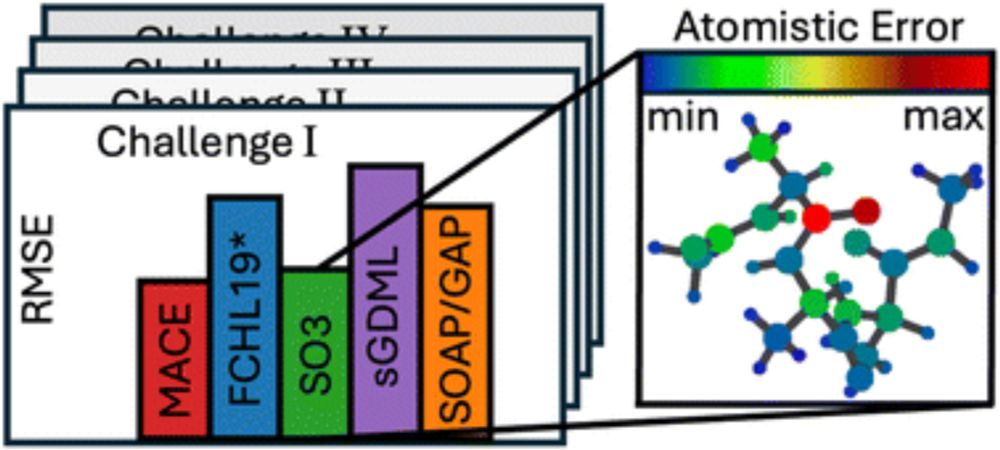
Link to our work to predict energy barriers from reactants without DFT: tinyurl.com/5cm4h3ka
@hitsters.bsky.social @mpip-mainz.mpg.de
08.04.2025 11:51 — 👍 14 🔁 4 💬 1 📌 0
A while online now already: Predict the barrier of a reaction without knowing the transition state and in the low data regime using Gaussian Process regr.
Led by Evgeni Ulanov, with Ghulam, Kai and Pascal Friederich @ KIT.
@mpip-mainz.mpg.de @hitsters.bsky.social
pubs.rsc.org/en/content/a...
31.03.2025 12:04 — 👍 15 🔁 4 💬 0 📌 0

NeurIPS participation in Europe
We seek to understand if there is interest in being able to attend NeurIPS in Europe, i.e. without travelling to San Diego, US. In the following, assume that it is possible to present accepted papers ...
Would you present your next NeurIPS paper in Europe instead of traveling to San Diego (US) if this was an option? Søren Hauberg (DTU) and I would love to hear the answer through this poll: (1/6)
30.03.2025 18:04 — 👍 280 🔁 160 💬 6 📌 12
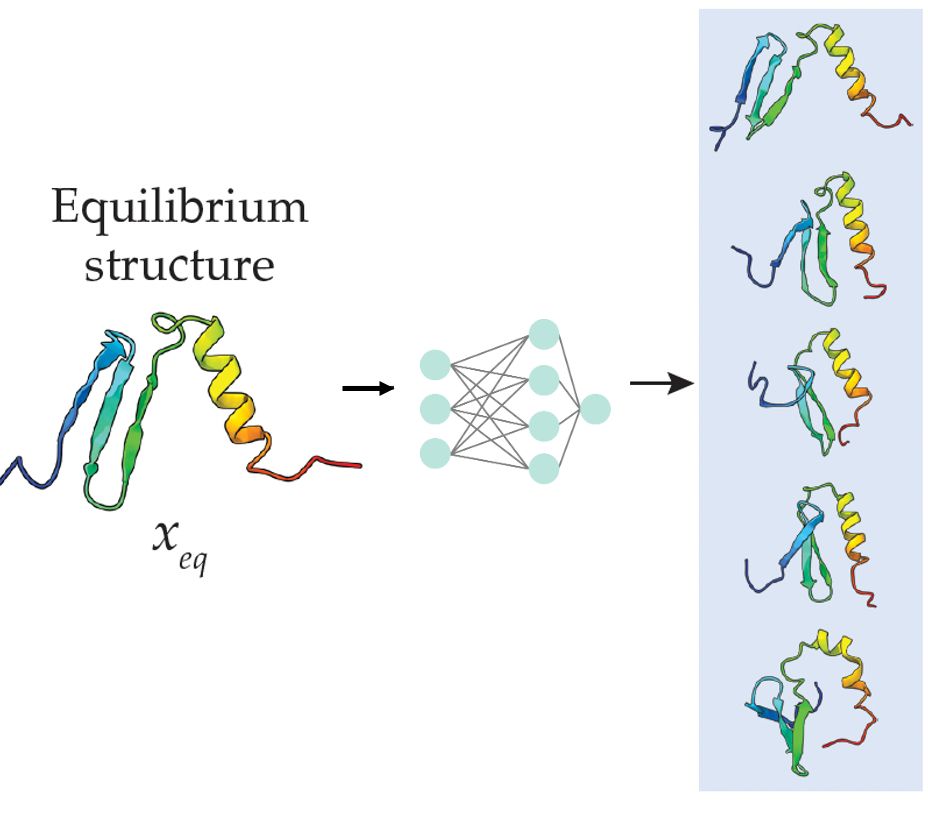
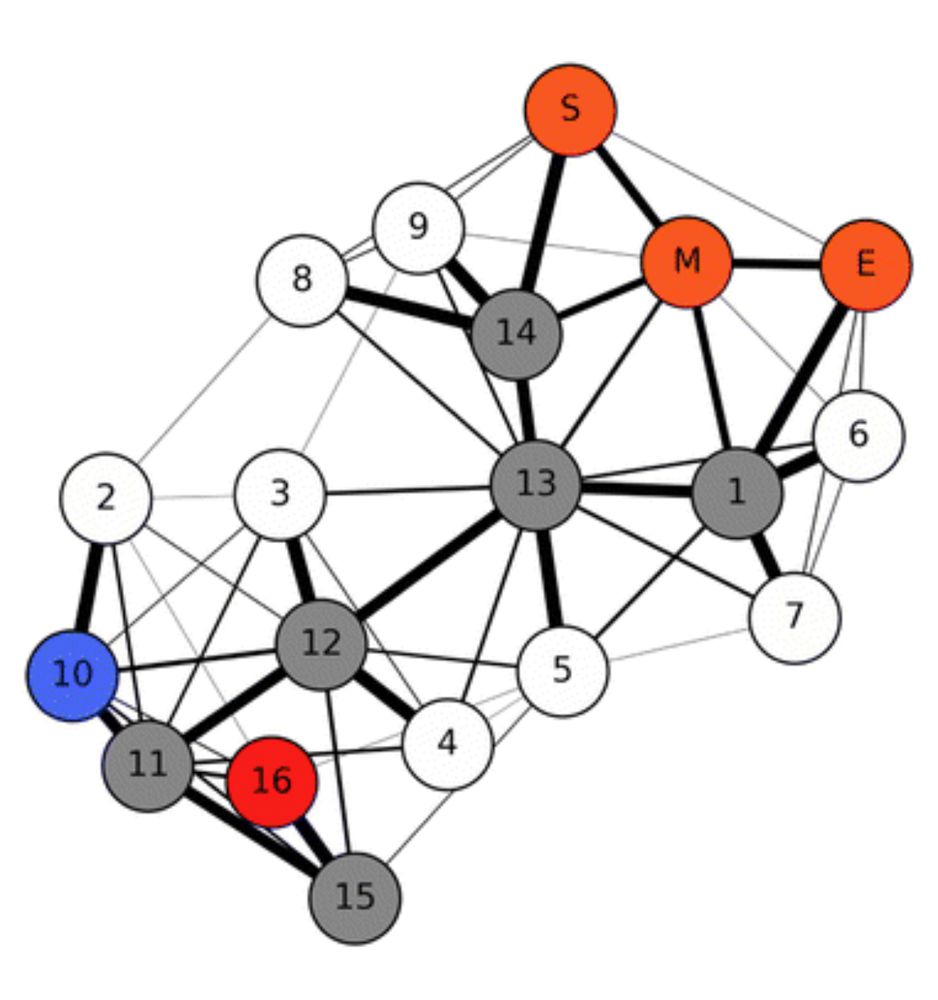
Wondering how to predict protein flexibility in a sec? No time to run MD simulations but want to go beyond pLDDT? Check out BBFlow
arxiv.org/html/2503.05...
Useful in particular for de novo designs.
Led by Nico Wolf & Leif Seute, w Seva, Simon, and Jan. @mpip-mainz.mpg.de @hitsters.bsky.social
31.03.2025 11:37 — 👍 39 🔁 9 💬 0 📌 0
YouTube video by TheHITSters
HITS-SIMPLAIX Joint Colloquium: Cecilia Clementi on protein dynamics and Machine Learning
Curious about how Machine Learning and Molecular Simulations are used to model Protein Dynamics?
Then watch our HITS-SIMPLAIX Joint Colloquium with Cecilia Clementi @cecclementi.bsky.social which is now available on our YouTube channel.
www.youtube.com/watch?v=2HAI...
12.02.2025 13:57 — 👍 4 🔁 3 💬 0 📌 0
#aufstehenfuerdemokratie
10.02.2025 08:33 — 👍 9 🔁 4 💬 0 📌 1
Don't miss our next HITS-SIMPLAIX Joint Colloquium on 10 February at 2:00 PM CET! Cecilia Clementi @cecclementi.bsky.social will talk about Modeling Protein Dynamics with Machine Learning and Molecular Simulation. Sign up ow.ly/h0Wl50UQ2kX to join via Zoom or just come by. @simplaix.bsky.social
03.02.2025 14:38 — 👍 21 🔁 8 💬 0 📌 0
Thanks! If you talk metal ions you will need to train it for such systems, we haven't. Others got in touch with us that use it for this purpose - happily so far I think ;-)
28.01.2025 22:16 — 👍 1 🔁 0 💬 1 📌 0
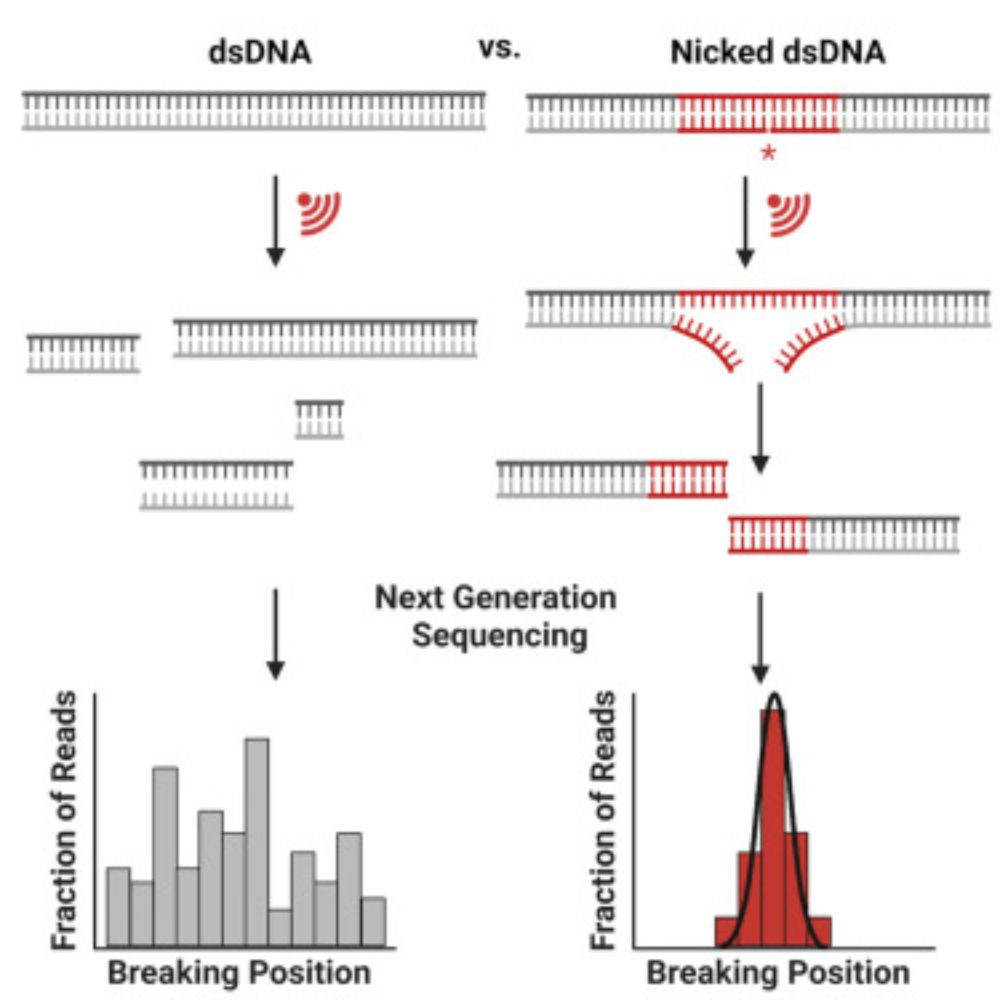
This was a very interesting paper to help supervise because it’s a totally different way to think about DNA structure, and uses a sequencer in a very creative way to understand DNA breakage. Check it out! 🧬
17.01.2025 10:19 — 👍 6 🔁 1 💬 0 📌 1
Need forcefield parameters for a weird small molecule, modified nucleotide, or non-natural amino acid? Check out Grappa, a GNN approach to generating bonded parameters compatible with the MD engine of your choice! 🧬
27.01.2025 16:41 — 👍 2 🔁 1 💬 0 📌 0

A long journey has taken the next step. Yesterday we defended our Cluster of Excellence Proposal #CoM2Life Communicating Biomaterials at the DFG. What an amazing journey with an amazing team. #teamscienceisthedreamscience @unimainz.bsky.social @tuda.bsky.social @mpip-mainz.mpg.de
15.01.2025 14:49 — 👍 17 🔁 3 💬 3 📌 1
🥲... and very excited to start from January with full forces at the MPIP@Mainz, with many (most) of my awesome lab members - and some HITS spirits - tagging along
20.12.2024 20:58 — 👍 6 🔁 1 💬 0 📌 0
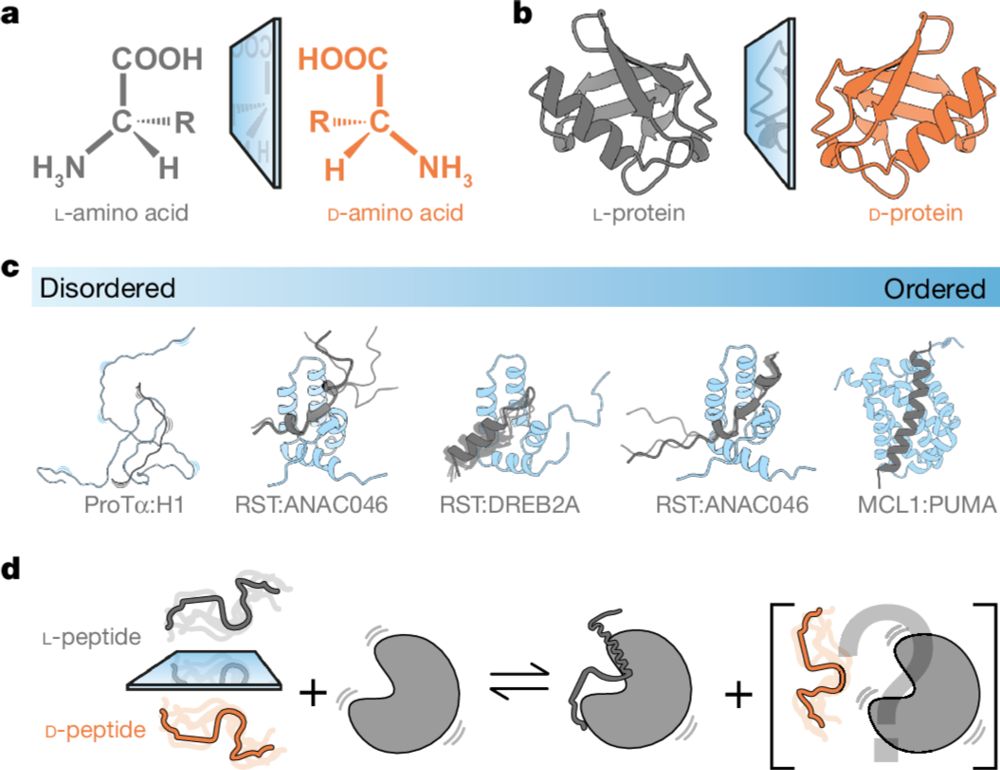
Stereochemistry in the disorder–order continuum of protein interactions
Nature - Studies on protein–protein interactions using proteins containing d- or l-amino acids show that stereoselectivity of binding varies with the degree of disorder within the complex.
Have you looked in the mirror recently? We took a good look at stereochemistry and how D- and L-ligands bind ... its all about disorder! Exited to share a true REPIN collaborative work led by @estellaan.bsky.social, A. Due and J. Olsen. Thanks for support NNF and DFF.
rdcu.be/d1F6q
28.11.2024 07:44 — 👍 38 🔁 13 💬 1 📌 1
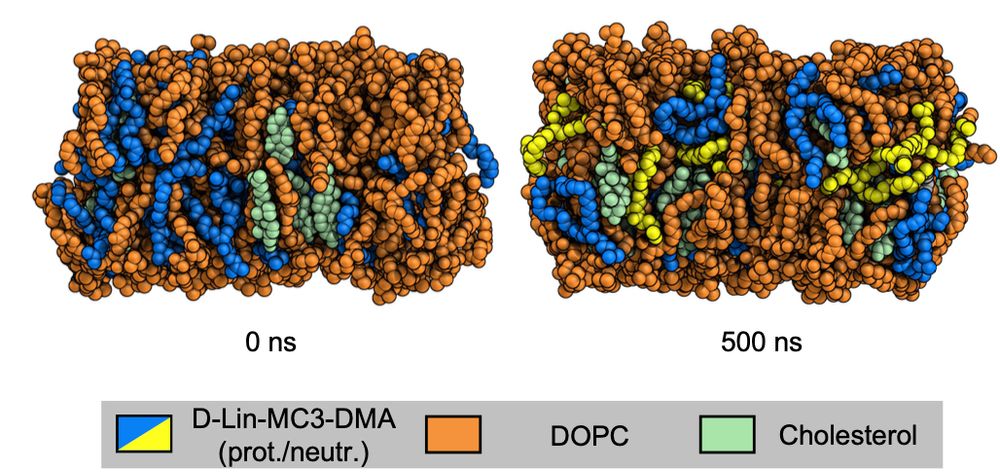
🚨 Preprint Alert! 🚨
Check out our new book chapter on Constant-pH MD Simulations of Lipids! Learn how to use CpHMD simulations to study pH-dependent behavior of aminolipids in lipid nanoparticles for drug delivery. 💊🧬
#membranes #LNP @m-a-r-i-u-s.bsky.social @fau.de
biorxiv.org/cgi/content/...
09.12.2024 20:37 — 👍 21 🔁 6 💬 1 📌 0
schurlab@ista.ac.at
Cryo-EM/Structural Biology/Cell Biology/Virology located @ ISTA
profile image credits: F-actin (Jesse Hansen/ISTA); photo (Anna Stöcher/ISTA)
Theory and computation of soft and condensed matter, dead or alive. Currently project leader at MPI of Biophysics in Frankfurt/Germany.
PhD candidate in bioinformatics
Protein structure prediction / Protein design
Posts by Division of Synthesis of Macromolecules @mpip-mainz.mpg.de | JACS Associate Editor
Striving for equality, diversity and representation in all aspects.
https://www.mpip-mainz.mpg.de/en/weil
Research group of Junior Professor Julia Westermayr
Wilhelm-Ostwald-Institute for Physical and Theoretical Chemistry
Leipzig University
Science Communicator, Head of Comms at the Heidelberg Institute for Theoretical Studies (HITS)
Molecular Simulations and Design Group.
Max Planck Institute for Dynamics of Complex Technical Systems, Magdeburg, Germany.
Prof. of Computational Chemistry
Heisenberg-Prof. (DFG)
Interdisciplinary Center for Scientific Computing
Heidelberg University
agamirjalayer.iwr.uni-Heidelberg.de
samirjalayer.de
Cooperation of @hitsters.bsky.social, @uniheidelberg.bsky.social & @kit.edu on bridging scales from molecules to molecular materials by multiscale simulation & machinelearning
Professor of Theoretical Chemistry @sorbonne-universite.fr & Director @lct-umr7616.bsky.social| Co-Founder & CSO @qubit-pharma.bsky.social | (My Views)
https://piquemalresearch.com | https://tinker-hp.org
Wir fördern Naturwissenschaften, Mathematik und Informatik.
#bildung #forschung #wisskomm
Datenschutz/Impressum: http://klaus-tschira-stiftung.de/datenschutz
Professor in bioinformatics, Stockholm University. Protein structure lover ( interactions predictions, evolution ..). Using machine learning as a part of AI for Sciences for halv my life. In addition to succén, I sometimes rants about sailing or skiing.
Professor at UC San Diego. Computational biophysics, biology, chemistry. Multiscale modeler. Viruses. Cancer. Co-Director of the Airborne Institute.
Associate Professor of Computational Chemistry, University of Birmingham
https://www.grynova-ccc.org/
views my own
Theoretical biophysicist who loves to talk about science.
Homepage https://www.thphys.uni-heidelberg.de/~biophys/
Scientist, #MachineLearning and #AI for Moleculear Sciences. Scuba Diver. Loves @cecclementi.bsky.social
Protein and coffee lover, father of two, professor of biophysics and sudo scientist at the Linderstrøm-Lang Centre for Protein Science, University of Copenhagen 🇩🇰
Professor at Stockholm University, Sweden. Weekend violinist. We study how enzymes work and how biological systems capture energy.