Global Analysis of Aggregation Determinants in Small Protein Domains
Protein aggregation is an obstacle for engineering effective recombinant proteins for biotechnology and therapeutic applications. Predicting protein aggregation propensity remains challenging due to t...
All data are available on Zenodo and we'd love to see what you can do with it! Cydney also made a nice notebook to run the predictive model (including DMS scan) in Colab!
Preprint: biorxiv.org/content/10.1...
Colab: colab.research.google.com/drive/1KNWvG...
Data: docs.google.com/forms/d/e/1F...
19.11.2025 21:16 — 👍 11 🔁 4 💬 1 📌 0
Exchanging for an authentic Wienerschnitzel 🤠
01.01.2026 12:12 — 👍 1 🔁 0 💬 0 📌 0


Biggest achievement of 2025 😂😂 First ever self-made cake seems to be a success!!
31.12.2025 20:37 — 👍 11 🔁 1 💬 1 📌 0
Are you interested in integrative structural biology, but feel a bit lost?
Don't worry, we have you covered with our FEBS Advanced Course, Lost in Integration Vol. 2 — probing biomolecules with AI and experiments
probingbiomolecules2026.febsevents.org
network.febs.org/posts/integr...
18.12.2025 19:27 — 👍 20 🔁 8 💬 0 📌 0
Folding models learn protein stability only implicitly. Without access to negative data, one can in principle make use of the folding free energy (dG) and the change in the free energy upon mutation (ddG). I believe simple aux losses could help for cases where a mutation is clearly disruptive.
18.12.2025 14:47 — 👍 0 🔁 0 💬 0 📌 0
Hi Thomas, that's indeed the case! However, this would also mean that one can reliably refold and score redesigned sequences only of those proteins whose structures were deposited post AF2 training cutoff date. This is a big limitation in our opinion!
17.12.2025 22:08 — 👍 0 🔁 0 💬 1 📌 0


Had a great time presenting our work on flexibility-conditioned protein structure design at @embl.org and right after our recent work on generating conformational ensembles of proteins at @euripsconf.bsky.social together with Nico. Follow along - we will be soon releasing more exciting things :)
05.12.2025 10:17 — 👍 5 🔁 0 💬 0 📌 0

New preprint out!
We present "Transferable Generative Models Bridge Femtosecond to Nanosecond Time-Step Molecular Dynamics,"
10.10.2025 13:45 — 👍 22 🔁 3 💬 1 📌 3

You asked and we listened... @workshopmlsb.bsky.social is excited to be expanding to Copenhagen, DK at @euripsconf.bsky.social 🎉
Two workshops (San Diego & Copenhagen) will run concurrently to support broader attendance. You can indicate your location preference(s) in the submission portal💫
12.09.2025 12:43 — 👍 10 🔁 6 💬 2 📌 2
Good luck and enjoy, Seb!
27.08.2025 08:45 — 👍 2 🔁 0 💬 0 📌 0

Hope you all had a good summer. I'm very happy to announce the speaker line-up for the falls Chalmers AI4Science seminars! Hope to catch you all there!
25.08.2025 10:41 — 👍 8 🔁 2 💬 0 📌 1
If you’re interested in learning more about protein folding and misfolding, I’ve created a convenient reading list with a few essential papers:
scholar.google.com/citations?us...
scholar.google.com/citations?us...
06.08.2025 19:01 — 👍 69 🔁 15 💬 4 📌 0
6/6 Amazing team work with great co-authors
@leif-seute.bsky.social, Nicolas Wolf, Simon Wagner, @bioinfo.se, Jan Stühmer and @graeterlab.bsky.social
Try out FliPS and BackFlip yourself, code and Google Colab tutorials are available on GitHub!
17.07.2025 04:27 — 👍 0 🔁 0 💬 0 📌 0
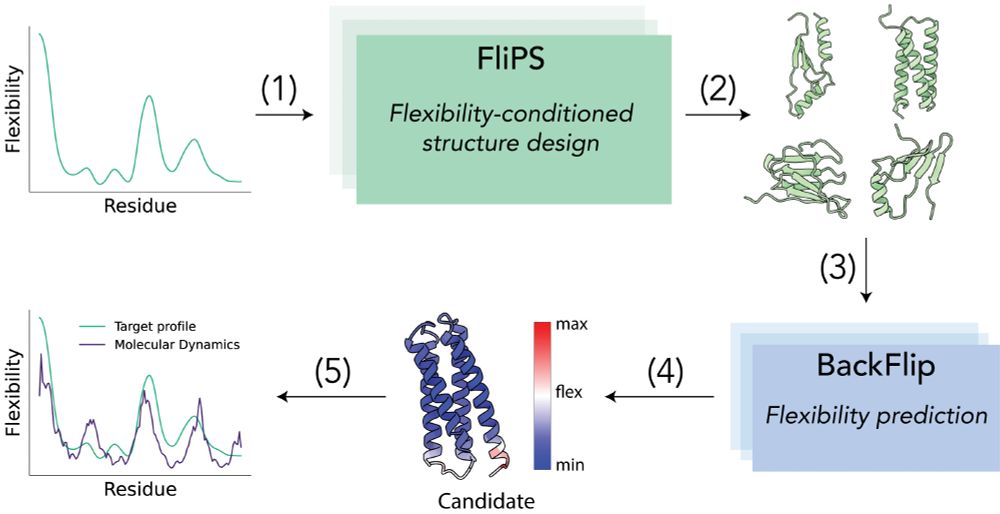
5/6 We introduce a framework in which we generate candidate protein structures conditioned on flexibility with FliPS and use BackFlip to select the candidates whose predicted flexibility profile best matches the target before running expensive MD simulations.
17.07.2025 04:21 — 👍 0 🔁 0 💬 1 📌 0
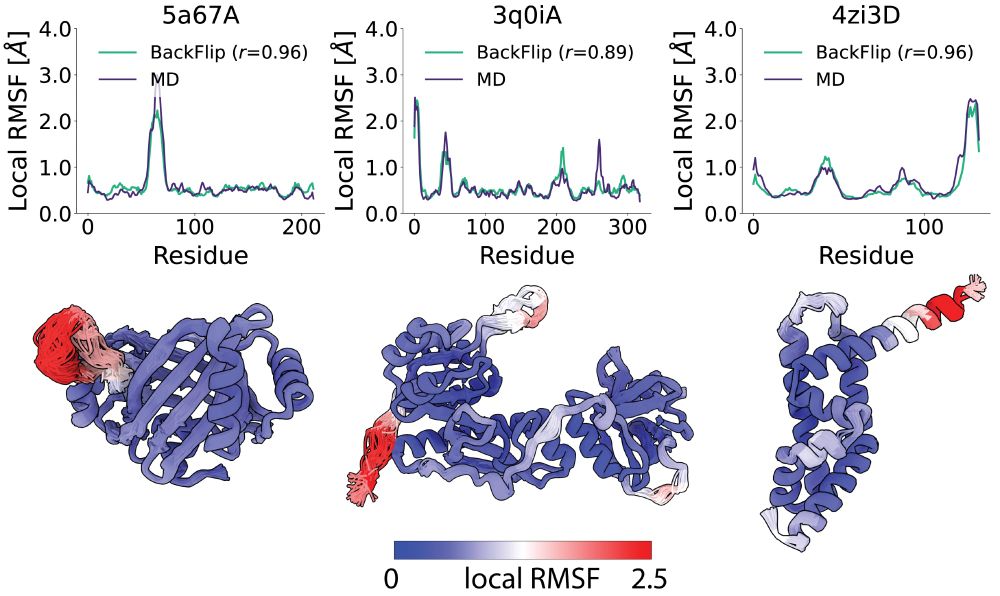
4/6 We also introduce BackFlip - an equivariant network that can accurately predict backbone flexibility as derived from MD simulations. Crucially, BackFlip infers flexibility solely from the backbone geometry without requiring evolutionary information, making it useful for de novo protein design.
17.07.2025 04:21 — 👍 1 🔁 0 💬 1 📌 0
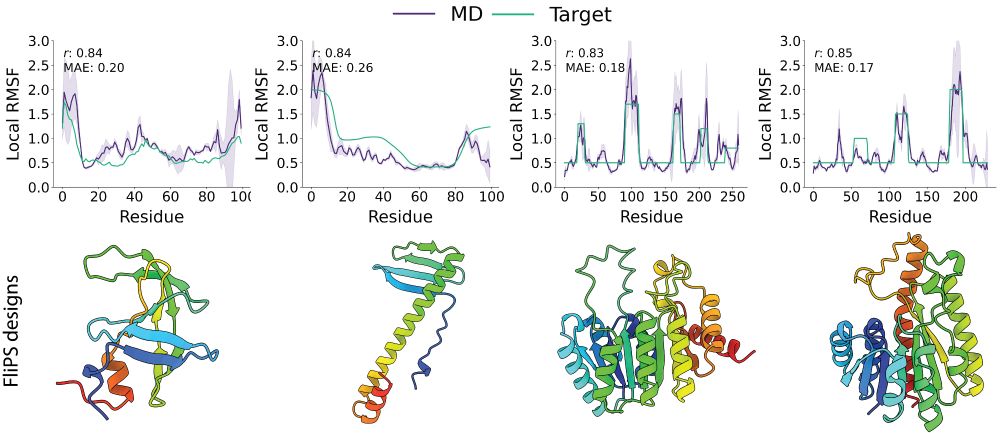
3/6 In a series of experiments, we demonstrate that FliPS samples novel, realistic proteins with diverse secondary structure composition and a remarkable resemblance to custom target flexibility profiles, as verified in 300ns Molecular Dynamics (MD) simulations of the designed samples.
17.07.2025 04:20 — 👍 0 🔁 0 💬 1 📌 0
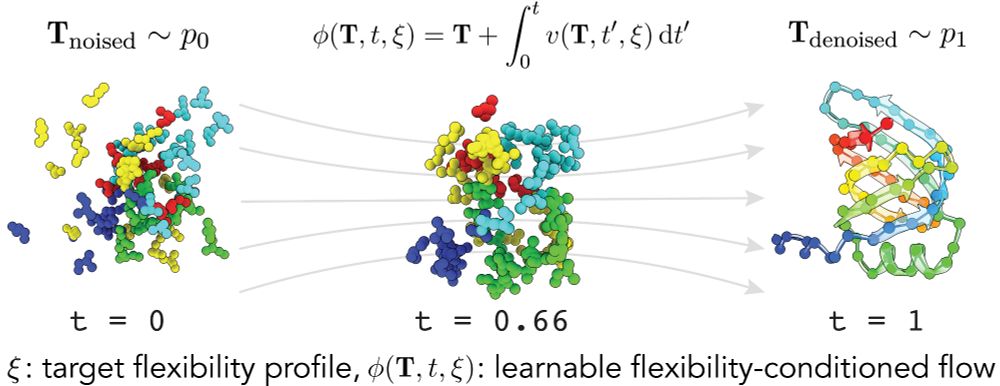
2/6 Our model FliPS is a conditional flow matching model for protein structure generation. FliPS receives a flexibility profile as conditional input feature and learns how to generate realistic protein structures while respecting target flexibilities.
17.07.2025 04:19 — 👍 0 🔁 0 💬 1 📌 0

1/6 One of the key features of functional proteins is their inherent structural flexibility. In our recent work at #ICML, we introduce flexibility to protein structure design! More in a thread below.
Code / Tutorial: github.com/graeter-grou...
Poster: W-109, Thu 17 Jul 11 a.m. PDT — 1:30 p.m. PDT
17.07.2025 04:18 — 👍 22 🔁 8 💬 1 📌 3


First time transatlantic and such a view over Greenland 🤯 Wish there were more glaciers :/
13.07.2025 21:57 — 👍 0 🔁 0 💬 0 📌 0
Hahaha this is hilarious!! I’m now totally convinced I should play with it as well 🤣
13.07.2025 04:52 — 👍 1 🔁 0 💬 0 📌 0

1/4
🚀 Announcing the 2025 Protein Engineering Tournament.
This year’s challenge: design PETase enzymes, which degrade the type of plastic in bottles. Can AI-guided protein design help solve the climate crisis? Let’s find out! ⬇️
#AIforBiology #ClimateTech #ProteinEngineering #OpenScience
08.07.2025 16:26 — 👍 23 🔁 20 💬 1 📌 4
This is not happening, right?
04.07.2025 07:10 — 👍 0 🔁 0 💬 0 📌 0
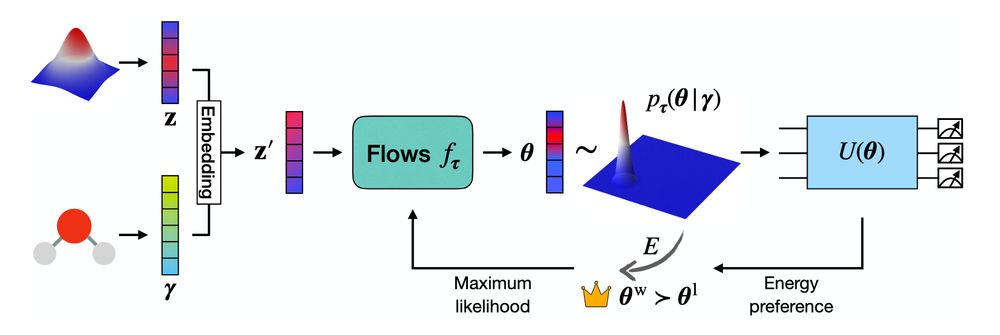
New pre-print from PhD student Hang Zou on warm-starting the variational quantum eigensolver using flows: Flow-VQE! Flow-VQE is parameter transfer on steroids: it learns how to solve a family of related problems, dramatically reducing the aggregate compute cost!
03.07.2025 08:18 — 👍 13 🔁 2 💬 1 📌 2
"...we investigated the performance of ... inverse folding models when trained on different structural datasets and on held-out set of experimentally determined PDB structures."
It would be interesting if they did it vice versa and ran inference on some FoldSeek or dark clusters of AFDB...
27.06.2025 12:36 — 👍 1 🔁 0 💬 1 📌 0
🫠
26.06.2025 11:40 — 👍 1 🔁 0 💬 0 📌 0
YouTube video by Science Communication Lab
The Most Beautiful Experiment: Meselson and Stahl
Just learned that Frank Stahl (of the Meselson and Stahl DNA replication experiment ("the most beautiful experiment in biology") died at the beginning of April, to no fanfare. Here's a lovely video of them reminiscing: www.youtube.com/watch?v=7-tn...
26.06.2025 10:01 — 👍 138 🔁 72 💬 5 📌 7
You can now check out a recording of Julija's excellent talk on the Chalmers AI4Science YouTube channel: www.youtube.com/watch?v=y_7a...
23.06.2025 07:03 — 👍 5 🔁 2 💬 0 📌 0
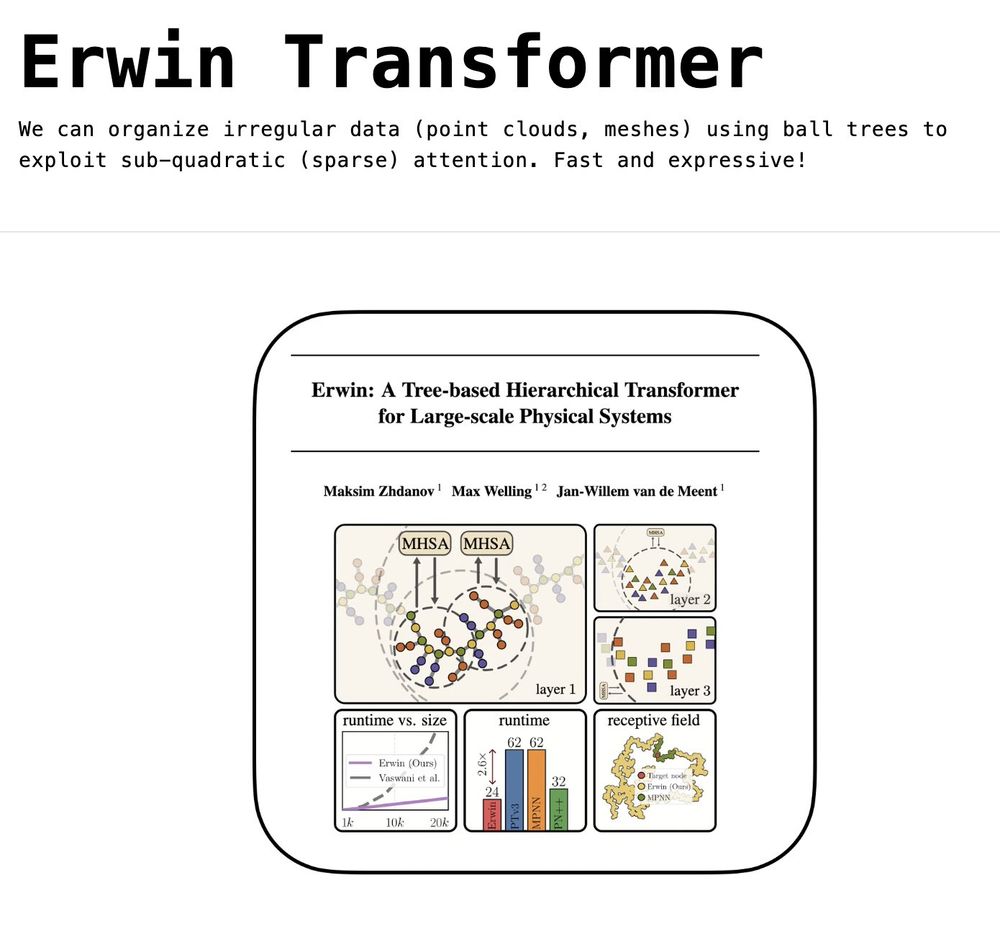
🤹 New blog post!
I write about our recent work on using hierarchical trees to enable sparse attention over irregular data (point clouds, meshes) - Erwin Transformer, accepted to ICML 2025
blog: maxxxzdn.github.io/blog/erwin/
paper: arxiv.org/abs/2502.17019
Compressed version in the thread below:
21.06.2025 16:22 — 👍 20 🔁 5 💬 1 📌 0
PhD Student in the Vorobieva Lab in de novo protein design
(he/him) Former PhD student / Postdoc @ Korbel group, EMBL.
Protein ML person, mathematics & science enthusiast.
developer of salad
preprint: https://www.biorxiv.org/content/10.1101/2025.01.31.635780v1
code: https://github.com/mjendrusch/salad
PhD student @mit.edu, previously @deshawresearch.com, @harvard.edu
The European Molecular Biology Laboratory drives visionary basic research and technology development in the life sciences. www.embl.org
PhD student at AstraZeneca and Chalmers University of Technology. Machine Learning for natural sciences (Drug discovery).
Computational biologist; Associate Prof. at University of Wisconsin-Madison; Jeanne M. Rowe Chair at Morgridge Institute
Biologist; Umass Chan Medical School
AITHYRA is a new dynamic research institute for biomedical AI in Vienna. AITHYRA seeks to build Europe’s premier institute for AI-driven biological and medical research, uniting computer scientists, engineers, and biologists in a collaborative environment.
Origins of life, astrobiology and synthetic biology group at Charles University in Prague
Machine Learning in Structural Biology workshop co-located with NeurIPS 2025 & at EurIPS 2025. Dec 7. San Diego, CA
http://mlsb.io
PhD student @univgroningen | Computational biophysicist | CG Martini
Assistant Professor at Stockholm University.
Dedicated Scientist.
🧪 Research Scientist @nvidia and PhD student @Oxford staring at proteins all-day
🧑💻 Website/Blog: https://kdidi.netlify.app/
🤖 GitHub: https://github.com/kierandidi
📚 Prev. Cambridge/Heidelberg
Improving life science with programmable experiments. 🧬 🤖 Home of the Protein Engineering Tournament & Open Datasets Initiative.
FutureHouse Fellowship - Sergey Ovchinnikov and Aaron Schmidt labs! Previously MIT EECS PhD with Debora Marks. Talk to me about modeling viral/immune proteins! 🦠
Prof @ Uni Groningen, metabolism, systems biology, @HeinemannLab, www.heinemannlab.eu
Trying to make this world a better place
Phase separation and disordered proteins!
All comments/opinions are my own and are not representative of St. Jude.