
Community perspective:
Toward a unified framework for determining conformational ensembles of disordered proteins 🍝
with framework for experimental data acquisition, computational ensemble generation & validation
Led by @hamidrgh.bsky.social, Silvio Tosatto & Alex Monzon
doi.org/10.1038/s415...
09.03.2026 18:58 —
👍 6
🔁 2
💬 0
📌 0
New paper from former PhD student @tkschulze.bsky.social on supervised learning of protein variant effects across large-scale mutagenesis datasets
MAVE/DMS experiments provide large amounts of data for benchmarking variant effect predictors, but may be difficult to use in supervised learning. 1/5
08.03.2026 08:40 —
👍 21
🔁 9
💬 1
📌 0
This turned into a rant by accident.
We are always thinking about this and are facing it right now: new molecules and some new routes to them with some new chemistry, some new Assays for the biology slashing costs by three orders of magnitude (not this research...but per sample moving forward)
08.03.2026 14:08 —
👍 0
🔁 1
💬 1
📌 0
We also see lab-to-lab variation despite using the same approaches (the top three panels are all from Doug's lab)
bsky.app/profile/lind...
As we discuss briefly in the paper, it should in principle be possible to learn the calibration curves from the MAVE data alone, but it's not easy
08.03.2026 12:46 —
👍 1
🔁 0
💬 0
📌 0
Genome Interpretation
Critical Assessment of Genome Interpretation
Thanks. One point of our paper was also to highlight that having calibration curves can be useful both for training and benchmarking, in particular when the latter occurs on a shared and physically meaningful scale.
As for tests on unseen data, I only know of CAGI genomeinterpretation.org
08.03.2026 11:19 —
👍 0
🔁 0
💬 1
📌 0

Review from Fia B. Larsen in @rhp-lab.bsky.social with everything you always wanted to know about proteasomal control of transcription factors (but were afraid to ask about)
Proteasomal control of transcription factors: mechanisms, regulation and dysregulation.
doi.org/10.1007/s000...
08.03.2026 10:34 —
👍 28
🔁 10
💬 1
📌 0
Supervised learning of protein variant effects across large-scale mutagenesis datasets
onlinelibrary.wiley.com/doi/10.1002/...
@tkschulze.bsky.social, Lasse Blaabjerg, @mcagiada.bsky.social
See also: Effects of residue substitutions on the cellular abundance of proteins
doi.org/10.7554/eLif...
5/5
08.03.2026 08:40 —
👍 7
🔁 1
💬 1
📌 0

Thea therefore built an approach to train models that takes this dataset-to-dataset variability into account via specific "standard curves", thus enabling training a model on the high-throughput data while learning to predict on the abundance scale only available in low-throughput experiments. 4/5
08.03.2026 08:40 —
👍 1
🔁 0
💬 1
📌 0
This variation makes it hard to perform supervised learning because a VAMP-seq score of, say 0.5, can mean quite different things in different datasets (see paper for a discussion of why that's the case). 3/5
08.03.2026 08:40 —
👍 1
🔁 0
💬 1
📌 0

Thea collected VAMP-seq data from the literature on how variants impact protein abundance, and showed that while there is a high correlation between abundance (as measured in low-throughput) and the sequence-based VAMP-seq scores, the relationship may be non-linear and vary across datasets. 2/5
08.03.2026 08:40 —
👍 1
🔁 0
💬 1
📌 1
New paper from former PhD student @tkschulze.bsky.social on supervised learning of protein variant effects across large-scale mutagenesis datasets
MAVE/DMS experiments provide large amounts of data for benchmarking variant effect predictors, but may be difficult to use in supervised learning. 1/5
08.03.2026 08:40 —
👍 21
🔁 9
💬 1
📌 0
That may be the situation in some cases but not in the one I refer to. I don’t think it would have made sense to publish method separately from this application. But strategically it might have been better. So in that case sort of the opposite of the (very real) issue to describe
07.03.2026 16:23 —
👍 2
🔁 0
💬 1
📌 0
I don't know. We tried to describe the new technique with a few equations in the main text as well as a flow-chart and benchmarking using synthetic data, targetting what I thought was the right the audience. And we did cut some corners compared to the full Bayesian approach.
07.03.2026 14:44 —
👍 1
🔁 0
💬 0
📌 0

Don’t hide the good stuff in the SI
In 2008, we published a paper on parameterizing force fields for unfolded proteins using NMR data, developing an early HPS model for intrinsically disordered proteins. The paper showed the idea, validation with synthetic data, and applications to real proteins.
07.03.2026 14:34 —
👍 12
🔁 3
💬 2
📌 0
FMLWY are wrong, so that’s indeed more than half that are correct if we accept the weird locations of the ring heteroatoms, how the OH’s are connected and the wrong names for DE
06.03.2026 22:01 —
👍 1
🔁 0
💬 3
📌 0

Design of the protein FRET ladder
Fancy a fresh preprint for Friday? When we were first getting involved with single molecule FRET, there weren't any standard protein molecules that suited our applications to help us develop our pipeline. So we built some! A universal protein ladder for FRET. 🧵 1/
06.03.2026 12:45 —
👍 43
🔁 13
💬 1
📌 2

The Division of Biological Physics @mpipks.bsky.social seeks a Research Group Leader in #biophysics, #softmatter physics, or related areas. (Further particulars in the ad.)
Apply by the 3rd of April 2026 at pks.mpg.de/bprgl to join us in Dresden!
03.03.2026 13:13 —
👍 17
🔁 8
💬 0
📌 1

10712 cups/12 years ~ 2.5 cups/day
Coffee and Tea Intake, Dementia Risk, and Cognitive Function
doi.org/10.1001/jama...
02.03.2026 21:22 —
👍 0
🔁 0
💬 1
📌 0

My trusted Jura has made myriad coffee and needed some TLC. Look forward to having it back
02.03.2026 20:53 —
👍 4
🔁 0
💬 1
📌 0

First preprint of the @pollyfordyce.bsky.social and @dunnlab.bsky.social collaboration! We used high-throughput microfluidics for sequence-strength mapping at the single-molecule level. Our new tech allowed us to discover a fundamental nonequilibrium property of multivalent systems. 1/13
27.02.2026 19:21 —
👍 15
🔁 8
💬 1
📌 1
Silvia's simulations showed that while all epitopes expose Sia for binding, Siglec-6 recognises and binds only GM1 because of a key interaction with the membrane through W127 and K126, which orientates the V-set domain to bind the Sia through Arg122 and the terminal Gal to the C-C' loop 😎
02.03.2026 17:59 —
👍 13
🔁 4
💬 1
📌 1
Maddipatla, Rzayev, Pegoraro, Pacesa, Schanda, Marx, Vedula, Bronstein: Inference-time optimization for experiment-grounded protein ensemble generation https://arxiv.org/abs/2602.24007 https://arxiv.org/pdf/2602.24007 https://arxiv.org/html/2602.24007
02.03.2026 06:49 —
👍 1
🔁 2
💬 0
📌 0
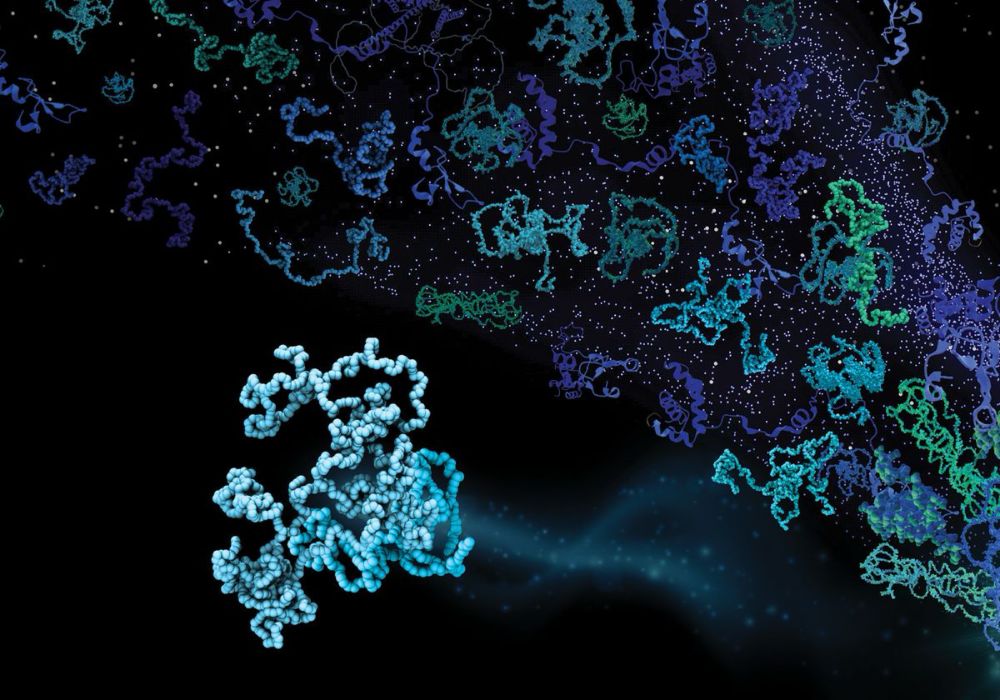
The Dynamic Lives of Intrinsically Disordered Proteins
Shapeshifting proteins challenge a long-standing maxim in biology.
Very nice article by Danielle Gerhard in The Scientist on
“The Dynamic Lives of Intrinsically Disordered Proteins”
with quotes from Gabi Heller, @alexholehouse.bsky.social and myself about our shared love of and fascination with these proteins
www.the-scientist.com/the-dynamic-...
16.09.2024 17:02 —
👍 24
🔁 12
💬 0
📌 0
RNA: simple alphabet, complex grammar
— Sandro Bottaro, 2017
The most consistent thing I have learnt about RNA is that my intuition based on proteins tends to fail when applied to RNA
25.02.2026 20:59 —
👍 15
🔁 1
💬 0
📌 0

President Ribbon
Whoo hoo!! What an honor and a pleasure to add a new ribbon at the #BPS2026 this morning!
@biophysicalsoc.bsky.social
24.02.2026 17:11 —
👍 28
🔁 2
💬 1
📌 0
Yeah, it would perhaps have been better; I don’t think we ever got around to implementing it as the rotamer library background is only a small part of the prior that we use
23.02.2026 18:13 —
👍 1
🔁 0
💬 0
📌 0



















