Figure showing that MAP65-3 increases the microtubule binding of the C-terminal domain of POK2.
Shu Yao Leong, Erik Schäffer @schaefferlab.bsky.social & team @zmbp-tuebingen.bsky.social discover that the tail domain of the kinesin-12 POK2 acts as a versatile interaction hub for microtubules, MAP65-3 & lipids.
journals.biologists.com/jcs/article/...
journals.biologists.com/jcs/article/...
14.10.2025 15:13 — 👍 8 🔁 5 💬 1 📌 0

The tail domain of the plant kinesin-12 POK2 is a versatile interaction hub
Summary: In vitro reconstitution experiments reveal that the tail domain of the kinesin-12 POK2 interacts with microtubules, MAP65-3 and lipids as a possible mechanism for division site deposition and...
Another study from the lab🔬🎉
Our paper, "The tail domain of the plant kinesin-12 POK2 is a versatile interaction hub" is now published in the Journal of Cell Science!
Check it out: doi.org/10.1242/jcs....
#PlantCellBiology #Kinesin #CellDivision #Microtubules #ZMBP
14.10.2025 08:40 — 👍 9 🔁 5 💬 0 📌 0

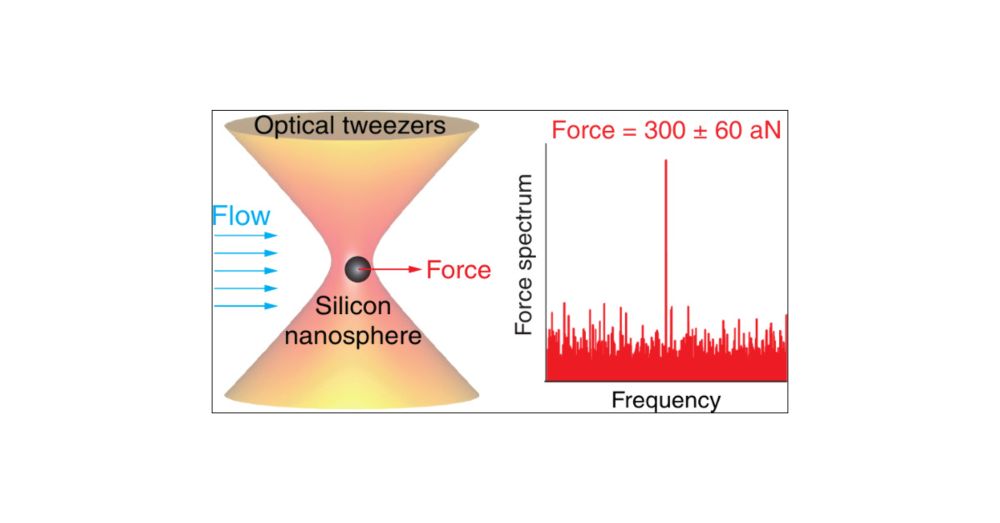
Exciting news! 🚀
Our latest work has been selected as the cover article in Nano Letters! 🎉
“Attonewton Force Resolution Measurements with Silicon Nanospheres at the Thermal Noise Limit in Ambient-Temperature Liquids”
#NanoLetters #CoverArticle #OpticalTweezers #ForceMeasurement #ZMBP
16.09.2025 10:47 — 👍 1 🔁 1 💬 0 📌 0


🎓 Huge congrats to Shu Yao on her successful PhD defense!
We're proud of her work and excited for what comes next! 🚀
#PhDDefence #graduation #cellularnanoscience #StructuralBiology #MolecularBiophysics #ZMBP #WomenInScience #AcademicMilestone
05.06.2025 13:32 — 👍 0 🔁 0 💬 0 📌 0
Congratulations to the #GreenRobust team for securing @dfg.de funding for the next 7 years to investigate plant robustness! 🌱
Special thanks to our very own Rosa Lozano-Durán @geminiteamlab.bsky.social and the co-spokespersons @thomasgreb.bsky.social and @kjschmid.bsky.social. Exciting times ahead!
23.05.2025 05:37 — 👍 50 🔁 15 💬 3 📌 1
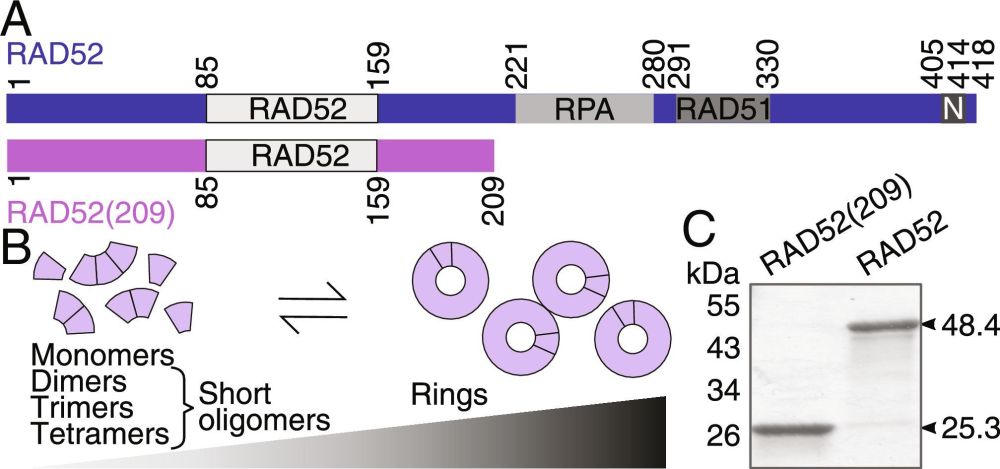
Monomers and short oligomers of human RAD52 promote single-strand annealing | PNAS
Genome maintenance and stability rely on the repair of DNA double-strand breaks. Breaks
can be repaired via the single-strand-annealing pathway med...
New paper alert! 📄
We’re thrilled to share a new publication from our lab:
"Monomers and short oligomers of human RAD52 promote single-strand annealing"
Check it out: www.pnas.org/doi/10.1073/...
#DNARepair #RAD52 #MolecularBiology #SingleMolecule #PhDResearch #MassPhotometry #StructuralBiology
08.04.2025 08:11 — 👍 5 🔁 2 💬 0 📌 0

Great time at #SMBioDresden — old friends, new friends, and exciting science!
#SingleMolecule #ScienceCommunity #AcademicLife #Dresden #MPI-CBG
19.03.2025 07:46 — 👍 5 🔁 1 💬 1 📌 0

Science runs on coffee, collaboration, and… Berliner! ☕🍩 Big thanks to Erik, for treating the team today. A great moment to recharge, share ideas, and enjoy the view from our lab!
#LabCulture #ScienceCommunity #coffeebreak
09.03.2025 11:19 — 👍 3 🔁 0 💬 0 📌 0
Journal of Cell Science (JCS) publishes cutting-edge science encompassing all aspects of cell biology. JCS is a community journal published by The Company of Biologists (@biologists.bsky.social), a not-for-profit organisation. #cellbiology #cellbio
RNA biologist, climate activist, and forest person. Director of Yale Center for RNA Science and Medicine.
🎥 https://boxd.it/1Ek8z
Biological algorithms of cell motility and pattern formation in active living matter, using theoretical physics https://physics-of-life.tu-dresden.de/friedrich
Professor of Physics (https://www.joerg-enderlein.de), Editor-in-Chief of Biophysical Reports (https://www.cell.com/biophysreports/home), Science & Arts, 🏳️🌈 | biophysics | single-molecule spectroscopy | super-resolution microscopy | nano-optics & plasmonics
Cryo-EM and cytoskeleton enthusiast. Professor of Structural Biology, Birkbeck College, London
We are a research lab in the Cluster of Excellence Physics of Life at the Technische Universität Dresden. Our interests are cell mechanics, cell biology and atomic force microscopy and modelling.
Theoretical physicist interested in the physics of living systems and statistical physics. Professor at LMU Munich. Passionate about emergent phenomena and interdisciplinary research.
https://www.theorie.physik.uni-muenchen.de/lsfrey/
Center for Plant Molecular Biology, University of Tübingen (Germany) supporting Independent Research Groups focusing on fundamental plant molecular biology since 1999
Biophysicist, senior researcher at Institut Jacques Monod, CNRS, université Paris cité
Regulation of cytoskeleton dynamics lab
single-molecule aficionado, antibiotic resistances
Prof at TUD
@tudresden.bsky.social Alum: UIUC, TUM
@tum.de, LMU @lmumuenchen.bsky.social, U Paris XI
Helping to understand life through the lens of single molecule biology at Exciting Instruments 🧬🔬
Formerly MPI-CBG and LUMICKS
The Nynke Dekker Lab at the University of Oxford
https://nynkedekkerlab.web.ox.ac.uk
single-molecule biophysics, biochemistry, DNA replication, chromatin, fluorescence, tweezers, and more!
The Ries group develops superresolution microscopy to study the structure and dynamics of proteins, ultimately in the living cell.
@maxperutzlabs.bsky.social @univie.ac.at
#SMLM, #MINFLUX, #endocytosis, #AI4microscopy
Theoretical biophysicist who loves to talk about science.
Homepage https://www.thphys.uni-heidelberg.de/~biophys/
Group Leader at the Azrieli Faculty of Medicine, Bar-Ilan University. Enthusiast of cilia, microtubules, motor proteins, and more.
https://www.orbachlab.com/
Account of the Max Planck Institute for the Physics of Complex Systems in Dresden; tweets by Pablo Pérez, Uta Gneisse, and Pierre Haas @lepuslapis.bsky.social.
A lab at the MRC LMB, Cambridge. Interested in dynein, microtubules, cytoskeleton, electron microscopy and much more.
Teaching, science, cytoskeleton, neurons, cilia, birds, gardens
Physics at Ulm University | live-cell/organism single-molecule microscopy | super-resolution microscopy | gene regulation | chromatin architecture | embryo development