The review also highlights exciting applications of SMT to nuclear factors and related findings, along with a perspective of future directions, including complementary genome-wide measurements of kinetic parameters.
I hope this review further sparks interest in and the application of live-cell SMT!
27.06.2025 15:40 — 👍 2 🔁 0 💬 0 📌 0
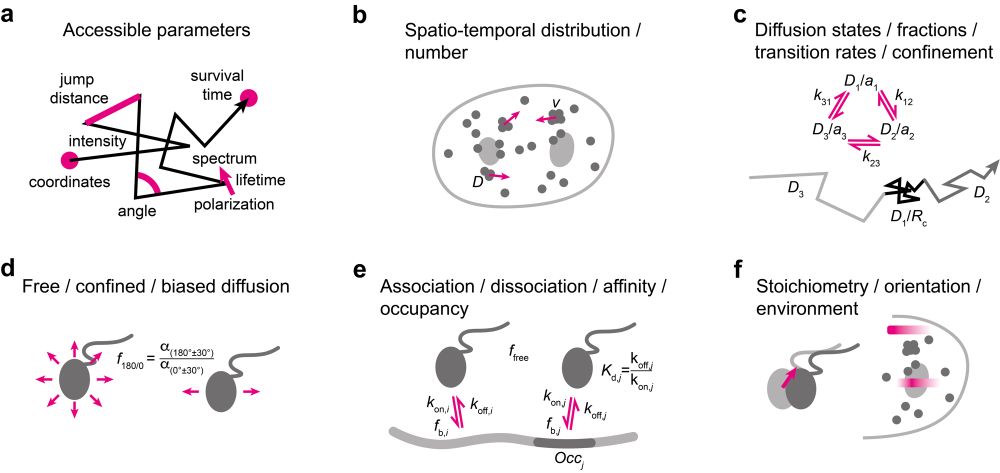
and a comprehensive discussion of kinetic parameters accessible by SMT.
27.06.2025 15:40 — 👍 3 🔁 0 💬 1 📌 0
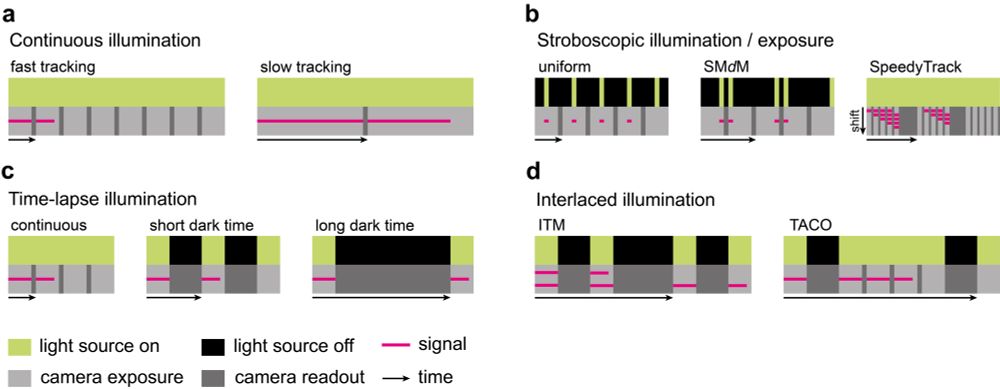
a discussion of temporal excitation patterns that facilitate measuring kinetic properties,
27.06.2025 15:40 — 👍 2 🔁 0 💬 1 📌 0
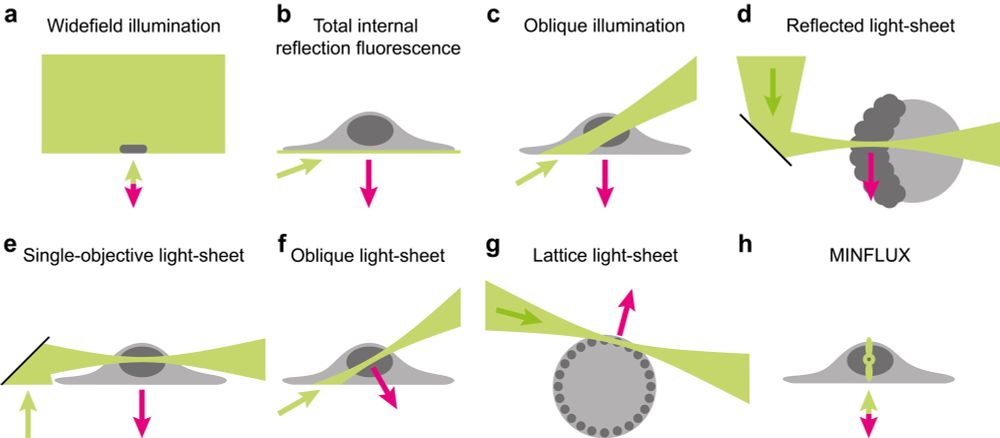
an overview of SMT microscopy techniques,
27.06.2025 15:40 — 👍 1 🔁 0 💬 1 📌 0
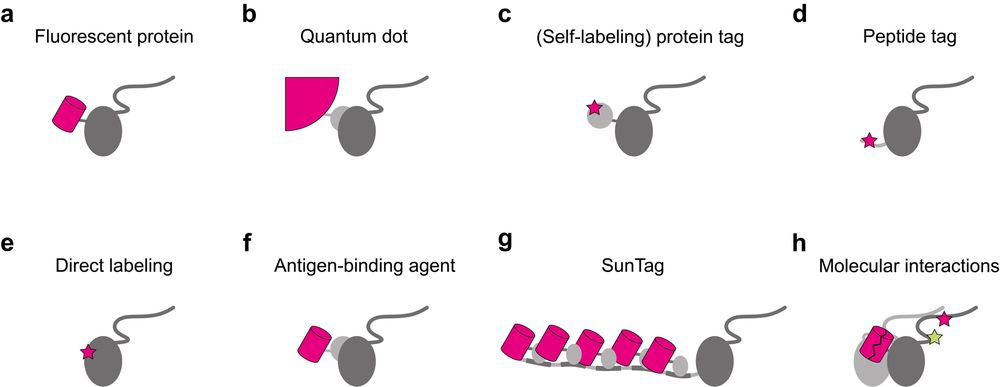
In the review, I give an overview of important steps along the implementation, execution, and analysis of single-molecule tracking in living cells and multicellular organisms. This includes a summary of labeling approaches,
27.06.2025 15:40 — 👍 1 🔁 0 💬 1 📌 0
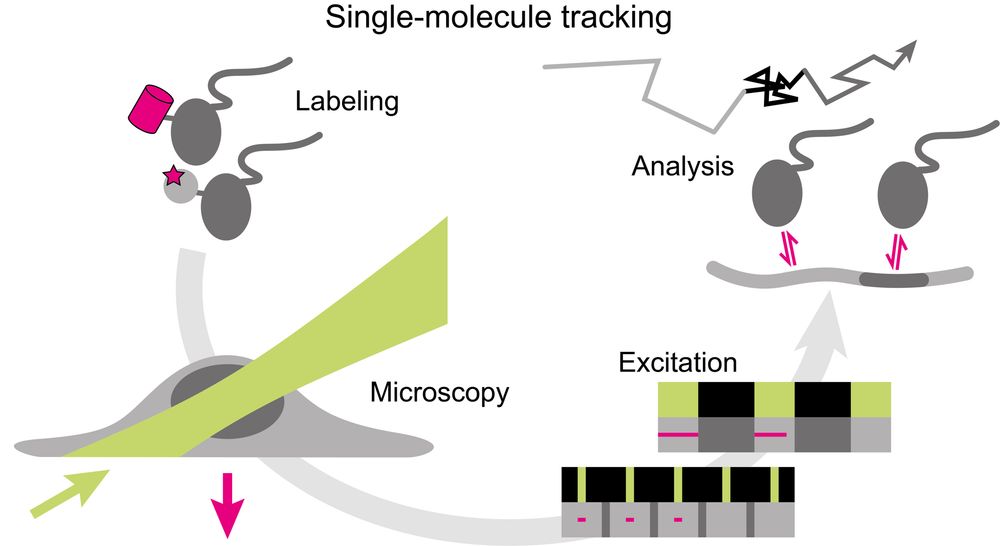
The review on live-cell SMT that I contribute to the jmolbiol.bsky.social special issue ‚Imaging of the central dogma‘ is now online as pre-proof: www.sciencedirect.com/science/arti...
27.06.2025 15:40 — 👍 11 🔁 3 💬 2 📌 2

This is a piece that I and @karsten-rippe.bsky.social discussing a lot, and a topic that is very close to my heart. The editors @naturerevgenet.bsky.social gave us the stage to do so, and the final version of our review is now available under this link: rdcu.be/erP1u
A short thread follows 1/n
19.06.2025 16:37 — 👍 95 🔁 39 💬 3 📌 2
So beautifully played :-) All the best for the final!
27.06.2025 03:32 — 👍 0 🔁 0 💬 1 📌 0
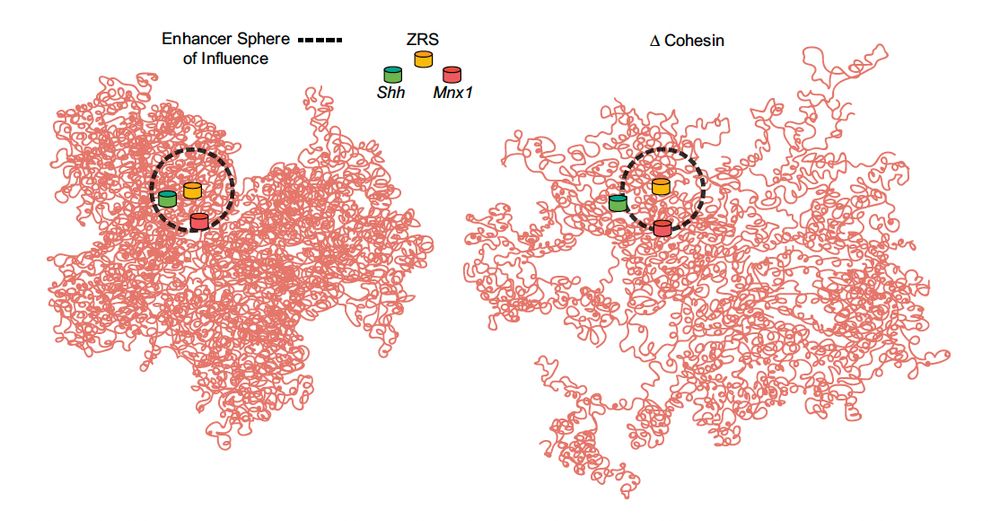
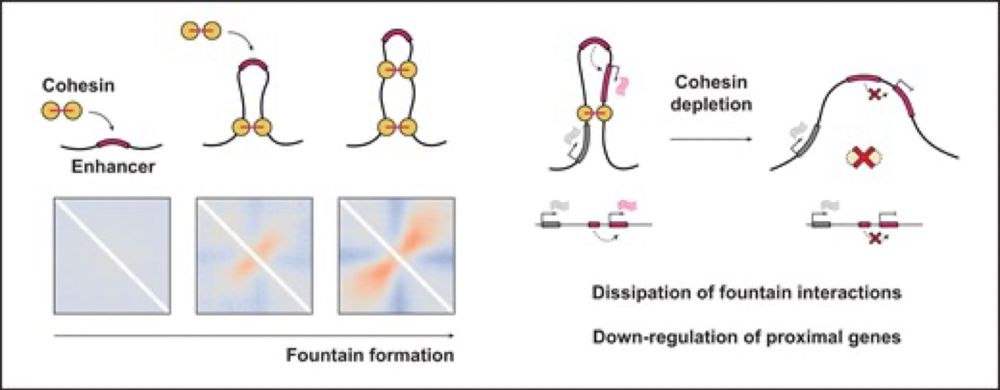
Great to see this published @genesdev.bsky.social. Evidence that enhancers can activate a bystander gene in an adjacent TAD, and that cohesin facilitates this. @uoe-igc.bsky.social.
pubmed.ncbi.nlm.nih.gov/40436628/
30.05.2025 14:13 — 👍 53 🔁 20 💬 1 📌 1
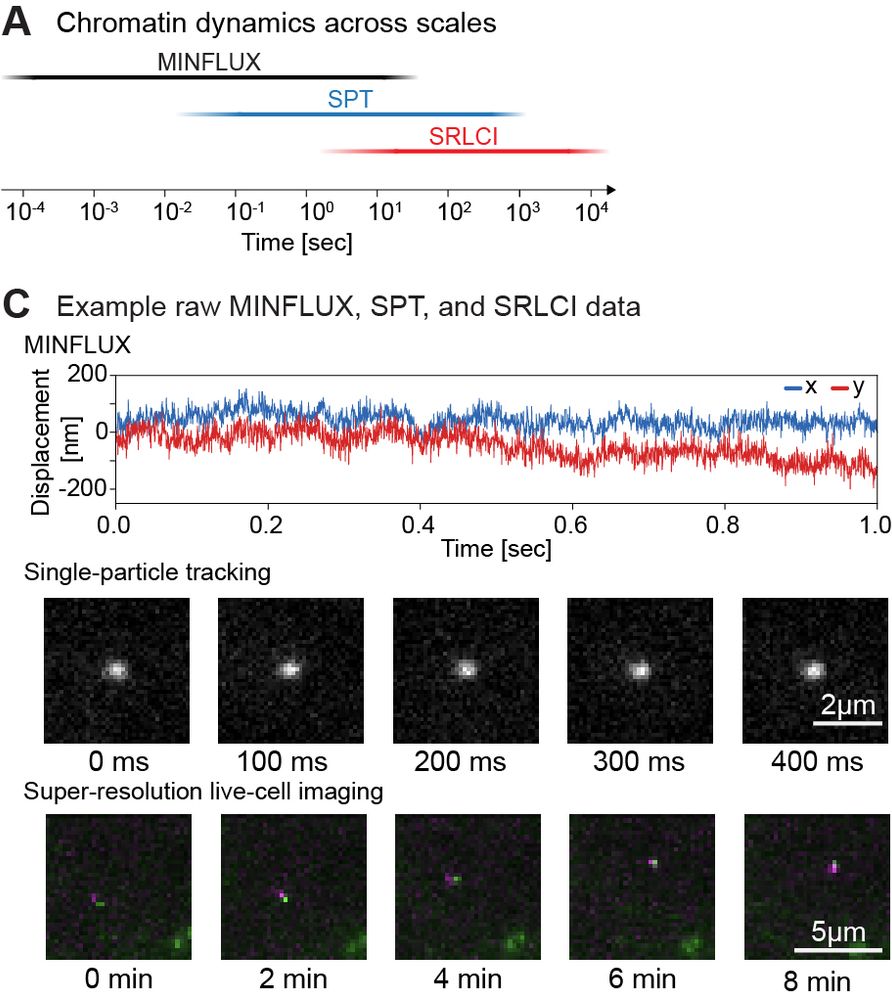
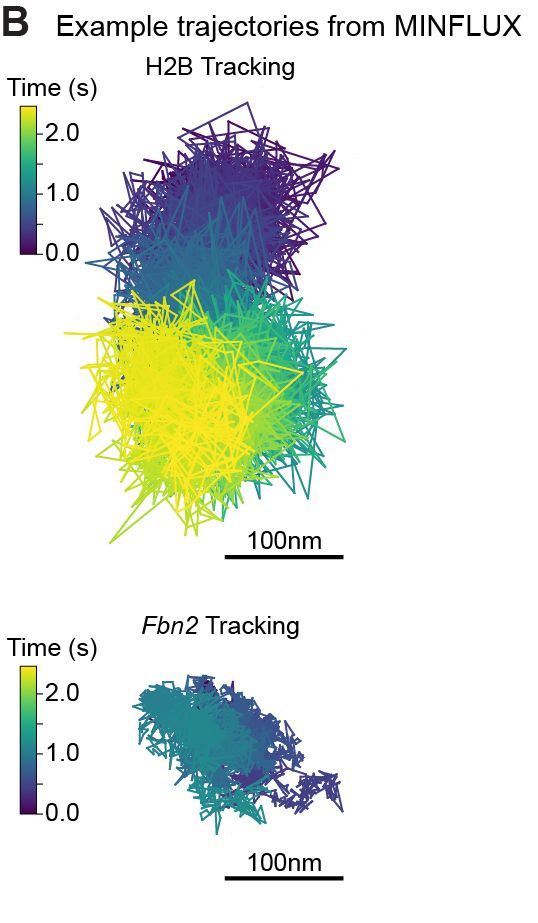
1/
Super excited to share my first preprint from @andersshansen.bsky.social Lab! We used MINFLUX to track chromatin (H2B-Halo and Fbn2 locus) at an unprecedented 200 μs, then combined it with SPT to span μs-minutes (H2B) or SPT & Super-Res Live-Cell Imaging (SRLCI) to span μs-hours (Fbn2)
14.05.2025 17:58 — 👍 29 🔁 11 💬 1 📌 4
📣 New preprint from the lab – Massive work entirely driven by super-talented @simsakong.bsky.social , jointly betw. @beatfierz.bsky.social lab and mine – combining in vitro and in vivo TF single molecule imaging
www.biorxiv.org/content/10.1...
12.03.2025 18:05 — 👍 18 🔁 7 💬 1 📌 1
Fun introduction!
12.03.2025 06:25 — 👍 0 🔁 0 💬 0 📌 0
Intriguing mechanism to balance production and degradation rates of proteins
12.03.2025 06:23 — 👍 1 🔁 0 💬 0 📌 0
Fantastic! Your playing is really great!
12.03.2025 06:14 — 👍 0 🔁 0 💬 1 📌 0

Gene regulation: one molecule at a time
Program is out for our first EMBL #singlemolecule gene regulation meeting! Check out the dedicated panel sessions on single molecule genomics, single molecule imaging and theory where we would like to brainstorm the future of the field:
Registration is open! 👇
www.embl.org/about/info/c...
21.02.2025 09:58 — 👍 12 🔁 9 💬 0 📌 0
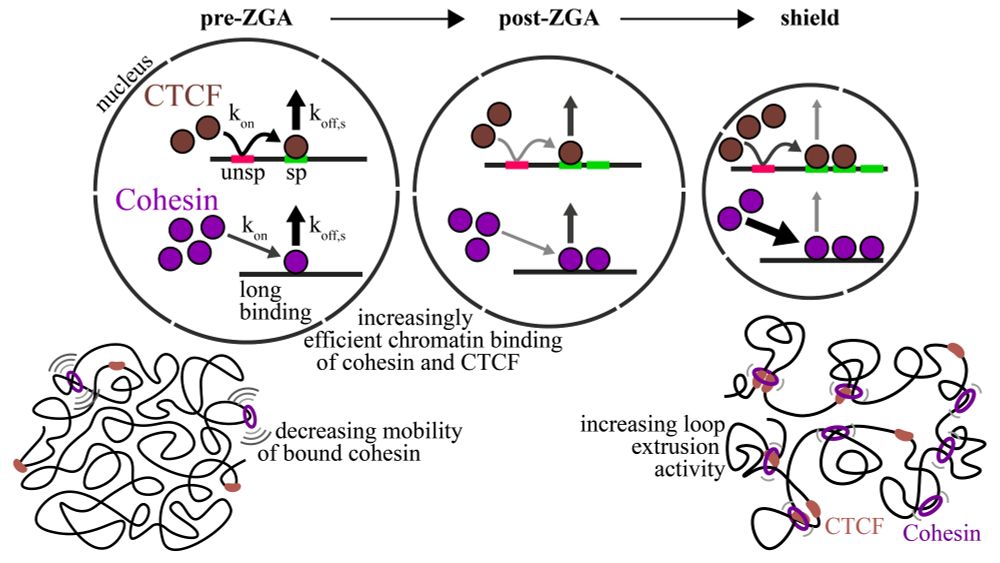
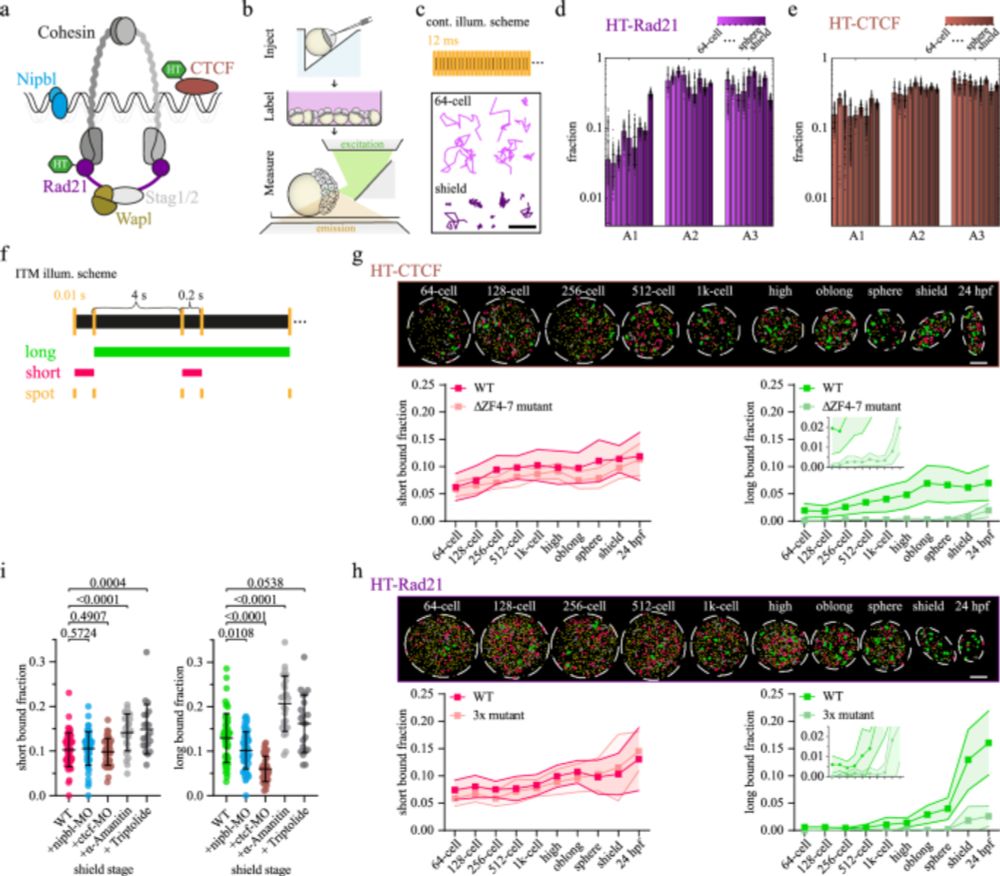
Search gets faster, binding longer, bound fraction of cohesin and CTCF goes up, especially at and after ZGA
21.02.2025 09:20 — 👍 4 🔁 1 💬 0 📌 0
#Cellpose 3 paper now out. Not all images are perfect. Restore your images with Cellpose3 to get better segmentations, w/ @marius10p.bsky.social www.nature.com/articles/s41...
12.02.2025 11:10 — 👍 137 🔁 49 💬 6 📌 2
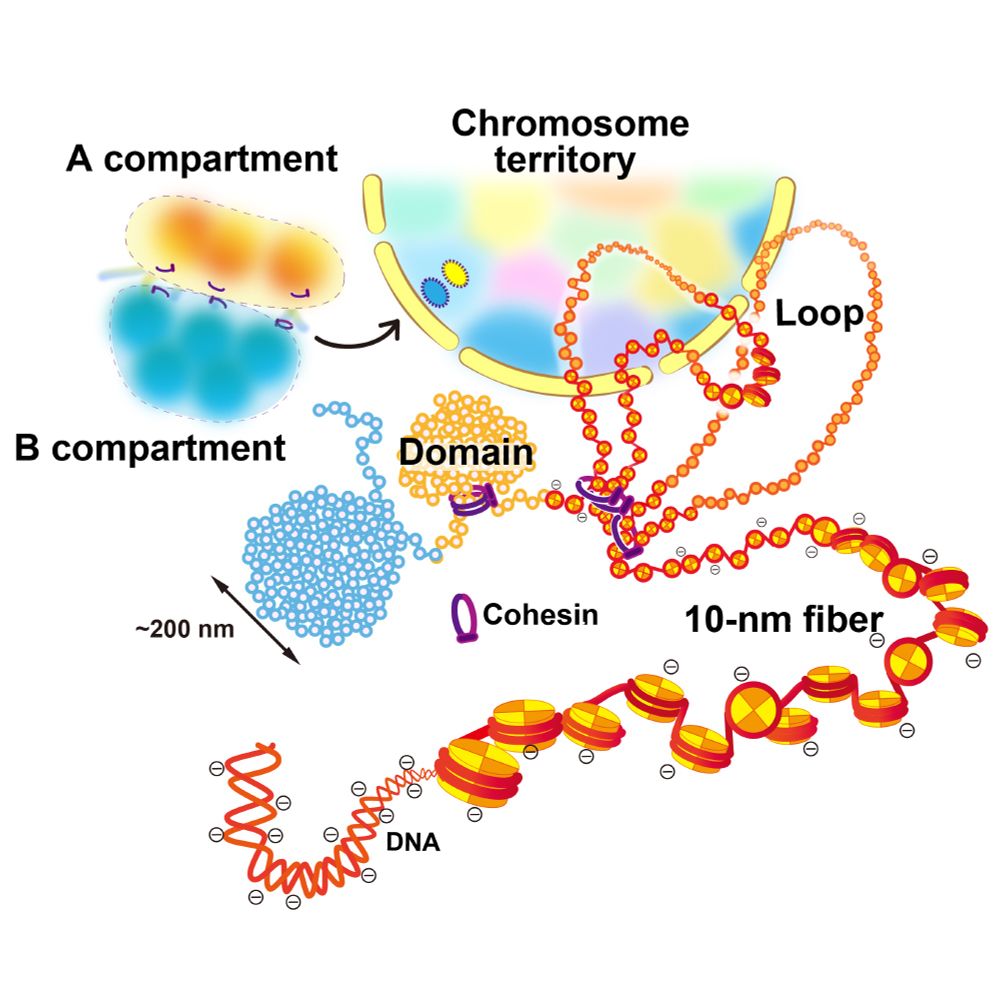
Our new review on how the #chromatin domain is formed in the cell is now available @Curr Opin Struct Biol.📄✨
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
20.02.2025 11:14 — 👍 74 🔁 36 💬 0 📌 0
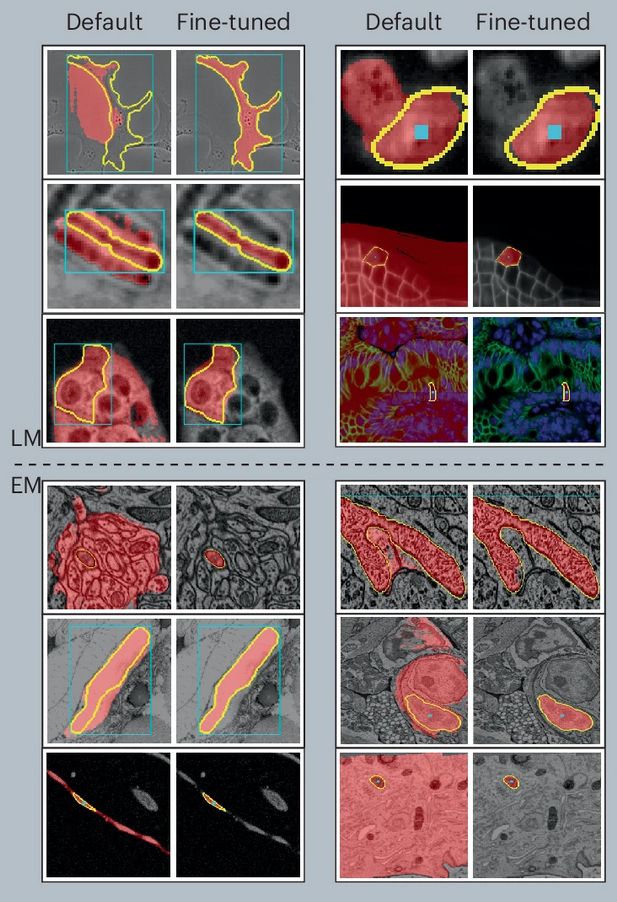
Improvements in LM (top) and EM (bottom) of our micro-sam model (finetuned) compared to the default SAM model.
After a long journey, Segment Anything for Microscopy is now published in Nature Methods! We significantly improve SAM for interactive and automatic segmentation in light and electron microscopy and build a user-friendly tool.
www.nature.com/articles/s41...
12.02.2025 11:41 — 👍 351 🔁 120 💬 11 📌 7
Fantastic work, @armelletollenaere.bsky.social , @davidsuter.bsky.social and co-workers! Happy we could contribute.
13.02.2025 15:34 — 👍 1 🔁 0 💬 0 📌 0
Big thanks to all people involved @armelletollenaere.bsky.social , Enes Ugur, Susanna Dalla-Longa, Cédric Deluz, Devin Assenheimer, @gebhardtlab.bsky.social , and Heinrich Leonhardt
And thanks ChatGPT for haikuing the abstract
13.02.2025 12:39 — 👍 2 🔁 1 💬 1 📌 0
1/n Excited to share our new preprint! Using live imaging in Drosophila embryos, we show that RNA Pol II clusters switch from sites of initiation to elongation during zygotic genome activation and that they are stably associated with an active gene: www.biorxiv.org/content/10.1...
12.02.2025 11:36 — 👍 100 🔁 32 💬 5 📌 4
Great work and very useful developments!
12.02.2025 07:25 — 👍 1 🔁 0 💬 1 📌 0
Measuring morphogen transport over multiple spatial scales in live zebrafish embryos https://www.biorxiv.org/content/10.1101/2025.02.06.636788v1
09.02.2025 01:33 — 👍 4 🔁 5 💬 0 📌 1
After not having been on Twitter for a year, I somehow missed that everyone had migrated already.
07.02.2025 19:33 — 👍 1 🔁 0 💬 0 📌 0
Thank you! Happy this finally made it.
07.02.2025 19:29 — 👍 0 🔁 0 💬 0 📌 0
 02.02.2025 11:04 — 👍 1 🔁 0 💬 0 📌 0
02.02.2025 11:04 — 👍 1 🔁 0 💬 0 📌 0
Group leader of Chromatin (Dys)function lab @MDC-BIMSB
Former Postdoc @Mundlos lab
PhD @ Schirmer lab
https://www.mdc-berlin.de/robson
Postdoc @andersshansen.bsky.social Lab @ MIT | PhD @davidemazza.bsky.social Lab @ Vita-Salute San Raffaele University | 3D genome, TFs, gene regulation
#Chromatin biologist/biophysicist @NIG & SOKENDAI in Japan.
Chromatin is very dynamic and flexible, but NOT regular!!! 🧪🧬🔬
My career and work: http://bit.ly/2CuF4L5
Lab HP: https://bit.ly/3F1a8nk
YouTube Seminar: https://bit.ly/4fYZiOj
group leader @ HHMIJanelia, #neuroscience + AI 🔬 #cellpose | diversity and open-science for better science | Ⓥ | she/her | https://mouseland.github.io
Group leader @ Uni Göttingen for Computational Cell Analytics. Working on AI for biology and medicine with a focus on microscopy image analysis with deep learning.
https://user.informatik.uni-goettingen.de/~pape41/
Getting to grips with chromosome organisation and dynamics using ‘scopes, sequencing and silicon.
Located at @imbavienna.bsky.social - @viennabiocenter.bsky.social
Exploring RNA & RBPs in cancer: from cell edges to the chromatin. Microscopy lover, proteomics newbie. Likes computer sciences, physics & photography. Also an illustrator and graphic designer for Science and Biology (see: @STEMDORADOscimag)
Scientist, molecular biologist. Working in Institut de Génétique Moléculaire de Montpellier (IGMM, CNRS, Montpellier University. Transcription, RNA biology, Genome stability. Running away from X.
Assistant Professor of Cell Biology at Harvard Medical School | HHMI Freeman Hrabowski Scholar | Nexus of chromatin, transcription, replication, and epigenetics. farnunglab.com
Chair of the Department of Biomedical Informatics at the University of Colorado School of Medicine. Research: transcriptomics, machine learning, public data - pick two of three. He/him. Views mine, not employer's.
An international online community and seminar series about chromatin and gene regulation.
Join the community on Discord: http://discord.gg/dXqT89r
Subscribe to our mailing list: https://stats.sender.net/forms/bmVp3d/view
Senior scientist @Princess Margaret Cancer Centre, Prof @UofT. BHAG: Conquer cancer by treating it as a disease of the chromatin. https://lupienlab.uhnresearch.ca/
Professor, Department of Biological Sciences, University of Pittsburgh. RNA polymerase II mechanism, classic films, #classiccountdown once in awhile
Gene regulatory networks and genome evolution. How do single cells make up their minds?
@NimwegenLab@mstdn.science
@NimwegenLab on twitter. Sorry X.















 02.02.2025 11:04 — 👍 1 🔁 0 💬 0 📌 0
02.02.2025 11:04 — 👍 1 🔁 0 💬 0 📌 0