Hey #TeamTomo …would you like to try some 🍦ICECREAM!? Our @unibas.ch colleagues over at the @ivandokmanic.bsky.social lab have developed a stunningly effective tool to de-noise and de-wedge your tomograms. The contrast and details will give you brain freeze! 🍨❄️🤯
Please try, we need feedback 🧪🧶🧬🔬
23.10.2025 17:57 — 👍 44 🔁 16 💬 1 📌 1
Amazing work, congratulations!!
16.10.2025 02:35 — 👍 0 🔁 0 💬 1 📌 0
Just some 3D classification 🤩 WOW.
Congratulations
16.10.2025 02:26 — 👍 1 🔁 0 💬 0 📌 0
Very excited that my final postdoctoral work is now online in Cell @cellpress.bsky.social.
www.sciencedirect.com/science/arti...
Using 16 (!) cryo-EM structures, we uncovered how the three proteins coronin, cofilin and AIP1 work together to rapidly disassemble actin filaments.
13.10.2025 09:04 — 👍 149 🔁 41 💬 6 📌 5
Wait uh, what?? 🤩
10.10.2025 00:56 — 👍 2 🔁 1 💬 0 📌 0

Check out our new preprint on an integrated pipeline combining in situ #cryo-ET with MALDI #MSImaging for single-cell identification and classification from previously analysed EM-grids.
Link: www.biorxiv.org/content/10.1...
19.09.2025 18:07 — 👍 54 🔁 14 💬 1 📌 2

Amyloid reconstructions from cryoSPARC
Helical Reconstruction of Amyloids in cryoSPARC?
We present guidelines, limitations & future perspectives for processing amyloids in cryoSPARC @structurabio.bsky.social.
Great collaboration of the Kelly Lab and @landerlab.bsky.social #cryoEM #amyloid www.biorxiv.org/content/10.1...
05.10.2025 04:16 — 👍 44 🔁 14 💬 3 📌 0
Muyuan Chen has turned structural biology into an immersive experience with his new video game Meowtabolism, now available on Steam.
Try the demo here: store.steampowered.com/app/4045010/...
Give Muyuan feedback: steamcommunity.com/app/4045010
#ScienceGaming #StructuralBiology #CryoEM #STEMOutreach
04.10.2025 13:34 — 👍 48 🔁 23 💬 1 📌 3
Initial attempt at replicating in relion (parameters in next post). This is for Aca2-RNA, using a 100k subset of the 2D-classified particles (no prior 3D cleanup).
A 1-class ab initio in relion, then local refinement in relion (1.8deg searches+blush) gives a nominally 3.3Å map; 3.5 Å w/out blush.
26.09.2025 02:28 — 👍 57 🔁 24 💬 1 📌 2
Time for a thread!🧵 How different is the molecular organization of thylakoids in “higher” plants🌱? To find out, we teamed up with @profmattjohnson.bsky.social to dive into spinach chloroplasts with #CryoET ❄️🔬. Curious? ..Read on!
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
25.09.2025 18:00 — 👍 137 🔁 44 💬 3 📌 6

Huntingtin binds and bundles F-actin!
Thrilled to team up with the Humbert (Sorbonne Université) and Song (KAIST) labs on this discovery.
We had a blast doing cryo-ET on this unexpected complex — and now so many new questions lie ahead!
#TeamTomo #CryoET #HTT #actin
doi.org/10.1126/scia...
25.09.2025 15:01 — 👍 44 🔁 14 💬 1 📌 0
Ever wondered how to position organelles in bacteria? Wonder no more!
Happy to have contributed to this fantastic story from @cellforganized.bsky.social' lab.
The avenues this opens in the synthetic biology space are enormous!
Movie: Carboxysomes positioned in E. coli!
Preprint: shorturl.at/D6xVg
23.09.2025 15:31 — 👍 32 🔁 15 💬 0 📌 1
A molecular-resolution look into the near-native architecture of the spinach chloroplast🌱. This one was a long time in the oven, but we're happy to finally share our "version of record". What long-standing debates did we settle? Check back for a short thread🧵 on Monday. #TeamTomo #PlantScience 🧪🧶🧬🔬🌾
20.09.2025 19:24 — 👍 137 🔁 46 💬 3 📌 0
Structural dynamics of mitochondrial ATP synthase in Chlamydomonas reinhardtii revealed by in situ CryoET https://www.biorxiv.org/content/10.1101/2025.09.10.674987v1
16.09.2025 16:30 — 👍 2 🔁 2 💬 0 📌 0
Impressive! Congratulations!
30.08.2025 15:18 — 👍 3 🔁 0 💬 0 📌 0
Wow, those membranes!
12.08.2025 19:16 — 👍 0 🔁 0 💬 0 📌 0

Research
Xu Lab of Computational Biology
New Title Alert: AITom- an open-source platform for AI driven cellular electron cryo-tomography analysis.
Learn more here: buff.ly/LLGhjvY
#SBGrid #SBGridSoftware #StructuralBiology
12.08.2025 16:03 — 👍 4 🔁 4 💬 0 📌 0
We found a new #ecological factor in #microbial mats: the #bacterial contractile injection system. Using #cryoET, #metagenomics, & quantitative #immunoTEM, we studied this feature directly in natural populations of bacterial #cells.
12.08.2025 11:50 — 👍 9 🔁 3 💬 0 📌 0
I knew Texas was big, but I didn’t know it encompasses 4 states
08.08.2025 14:31 — 👍 0 🔁 0 💬 0 📌 0
Lovely.
So many US states I never heard of before, I definitely have to study an US map much better
08.08.2025 06:34 — 👍 1 🔁 0 💬 0 📌 0
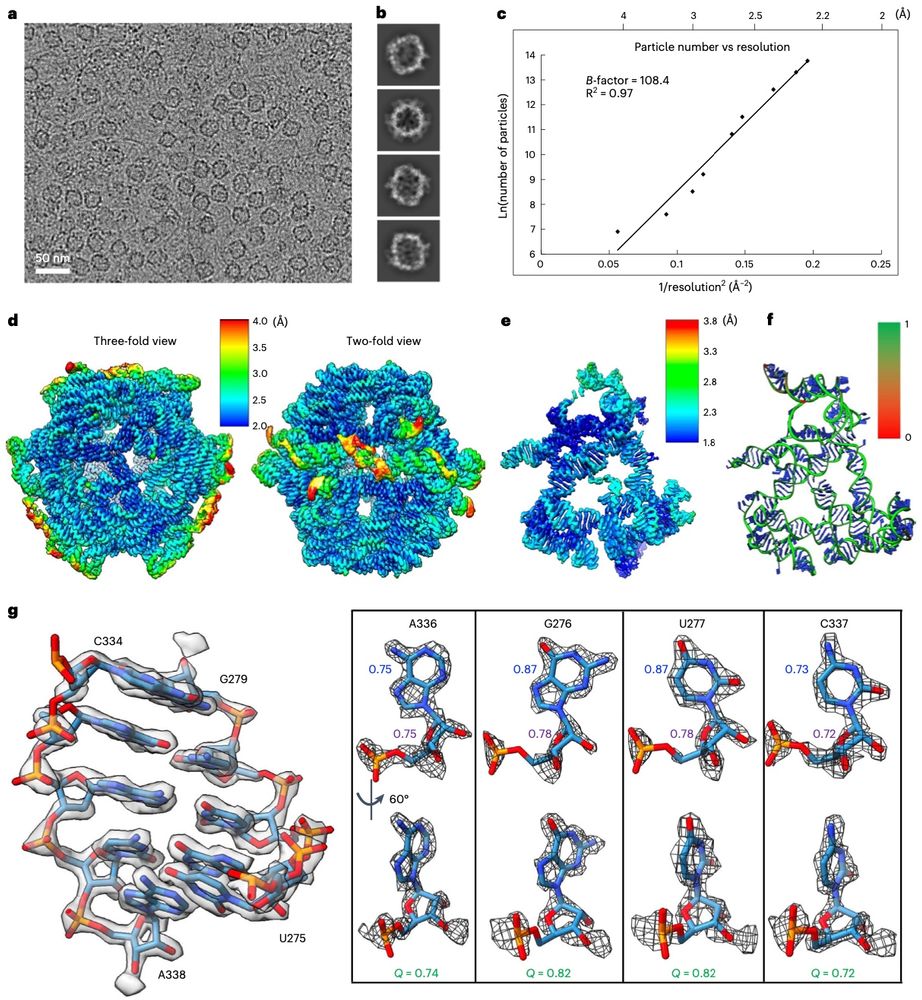
High resolution CryoEM maps of RNA assemblies
Not my area at all, but how cool are these cryoEM structures of purely RNA-based assemblies!
www.nature.com/articles/s41...
06.08.2025 22:41 — 👍 43 🔁 6 💬 0 📌 3
Cryo-EM structures of Egl-BicD-RNA complexes reveal how diverse mRNAs are selected for subcellular localization www.biorxiv.org/content/10.1101/2025.08.02.668268v1 #cryoEM
03.08.2025 18:40 — 👍 2 🔁 2 💬 0 📌 0
Check out our new preprint on the discovery of a molecular switch in NAC that mediates nascent chain sorting on the ribosome and prevents mitochondrial protein mistargeting by SRP. A great collaboration with the Shan Lab @Caltech and the Qi Lab @UVA: www.biorxiv.org/content/10.1...
01.08.2025 15:27 — 👍 82 🔁 28 💬 4 📌 2
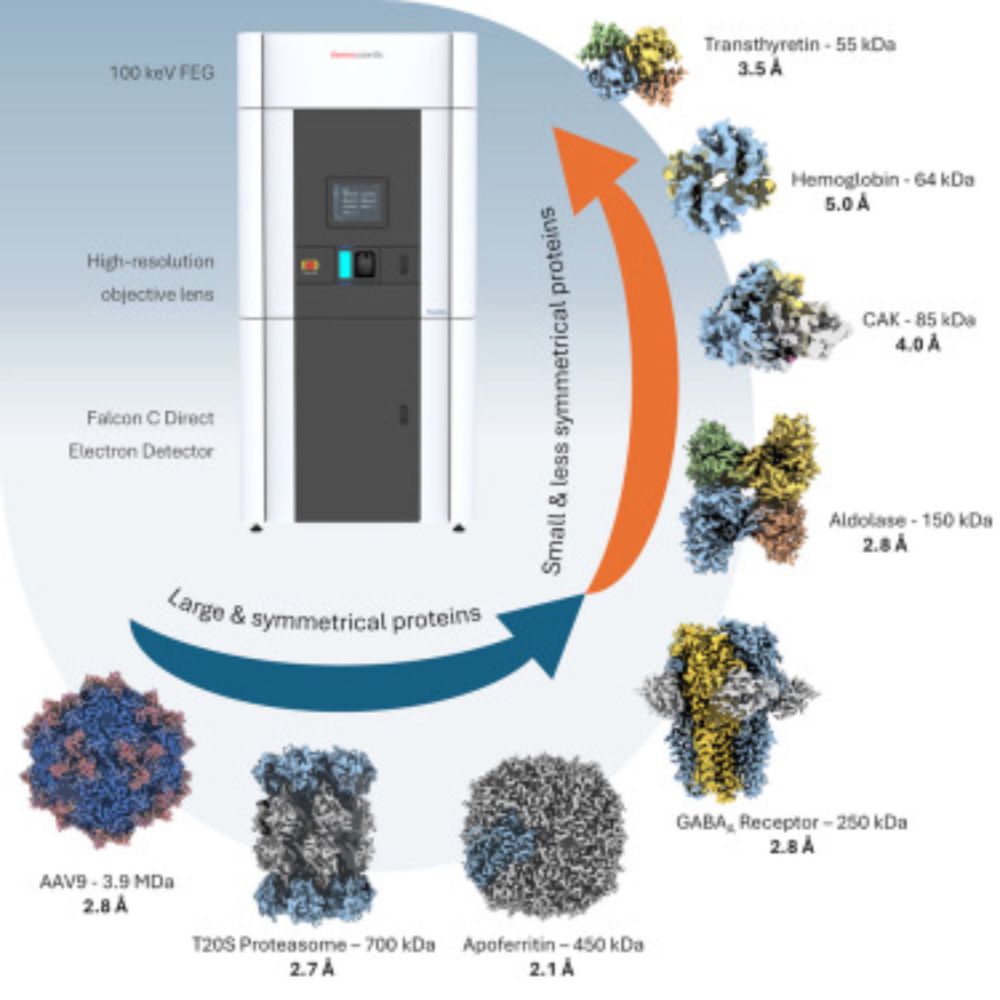
Sub-3 Å resolution protein structure determination by single-particle cryo-EM at 100 keV
Cryoelectron microscopy (cryo-EM) has transformed structural biology by providing high-resolution insights into biological macromolecules. We report s…
Now out in Structure: Paper on high-resolution #cryoEM using the 100 kV Tundra cryo-TEM by Dimple Karia, Adrian Koh, @abhaykot.bsky.social, @lingboyu.bsky.social, @landerlab.bsky.social and many others. Happy to have contributed to this effort. Check it out:
www.sciencedirect.com/science/arti...
30.07.2025 21:50 — 👍 80 🔁 37 💬 1 📌 0
I turn Art into Sports (and vice versa) | NO AI USED | “Everything I didn't know I needed" - follower testimonial
News from the Sternberg lab at Columbia University, Howard Hughes Medical Institute.
Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
Visit us at www.sternberglab.org
structural cell biologist @EMBL 🇨🇦
Structural biology, biophysics and microtubules @ ETH Zurich
Understanding and protecting the planet since 1903.
📍UC San Diego
Incoming Assistant Professor, Columbia University | @DamonRunyon.org Postdoc, UC Berkeley | PhD Princeton University | Mobile elements, Structural biology and Genome engineering | http://thawanilab.org
Postdoc in the Pauli Group @IMP in Vienna. Interested in gene expression regulation during early embryonic development in 🐟
America’s Finest News Source. A @globaltetrahedron.bsky.social subsidiary.
Get the paper delivered to your door: membership.theonion.com
I do social media workshops for scientists (Bluesky, LinkedIn, X, Reddit) and like talking #philosophy and #history when I am off duty! Ex-University Post editor in Copenhagen.
Website: https://mikeyoungacademy.dk/
Linktree: https://linktr.ee/mkeyoung
Wildlife, trees, terrestrial ecology, night, trees, corvids, coffee, cats. | Nocturnal (DSPD). AuDHD. They/them.
My nonhuman animal equivalent is the badger.
"Night is a world lit by itself."
—Antonio Porchi
Classical guitarist, recording artist, music educator, musicologist (PhD), Classical/Pop Int. Examiner 🎸💿🔎🎶
Follow me on 👇
YouTube: youtube.com/@FrancescoTeopini
IG: instagram.com/francesco_teopini
Spotify: https://open.spotify.com/artist/3mOYZ1urMzWb
Economic historian @UoGuelph w broad social science & historical interests: population health, First Nations demography, mobility, inequality & lives of the incarcerated 🇨🇦🇦🇺🇳🇿🏴
Editing Asia-Pacific Econ History Rev & directing https://thecanadianpeoples.com.
Professor passionate about student success & team science #firstgen #Brassica enthusiast walking Dogs of the Plant World
#Agriculture #Food #HigherEd #EduSky #Agsky #SciComm & more
#Soil #Crop #Science ; opinions mine
Demography nerd at Pew Research Center
Global religious change, sociology
Data scientist, data journalist, I like all things related to science, programming, discussions, societies, writing. Danish webpage with samples of work: tekstogtal.dk
Science integrity consultant and crowdfunded volunteer, PhD.
Ex-Stanford University. Maddox Prize/Einstein F Award winner
NL/USA/SFO.
#ImageForensics
@MicrobiomDigest on X.
Blog: ScienceIntegrityDigest.com
Support me: https://www.patreon.com/elisabethbik
climate scientist
posts 100% my own
🇨🇦 is my home
distinguished professor & chair, Texas Tech
chief scientist, The Nature Conservancy
board member, Smithsonian NMNH
alum, UToronto and UIUC
author, Saving Us
ITE assistant prof 👩🎓
Primary teacher EYFS to Y6 👩🏫
Secondary (physics) 💫
Engineer👩⚕️
Well-being of trainee teachers 🧚🏻♂️
First generation at uni 🙋🏼♀️
Anything physics, but love space🔭
mum👨👩👧👦
views mine, always learning















