Stay tuned for details on the 6th edition of MLSB, officially happening this December in downtown San Diego, CA!
28.07.2025 15:41 — 👍 14 🔁 7 💬 0 📌 0

Unfortunately, the MLSB Workshop @ NeurIPS (@workshopmlsb.bsky.social) was rejected this year.
Feedback from the deciding committee indicates it was a coin flip decision, with 283 proposals & a number related to “computational biology”
More on the future of MLSB soon…
07.07.2025 14:21 — 👍 8 🔁 5 💬 2 📌 2
Exactly. Also for some countries like Germany where people have been complaining about the conditions for research for years (people finishing their thesis with unemployment money and other shenanigans) it's kinda a slap in the face that suddenly there is money to bring in new hires...
05.05.2025 05:22 — 👍 1 🔁 0 💬 0 📌 0
Bioicons - high quality science illustrations
Submit new Bioicons under permissive CC0, CC BY or MIT License.
Not as pretty but this should give you a vector you can tune easily in a similar way with reduced color complexity (might be a bit slow for large proteins in contour mode) bioicons.com/pdb2vector/ And uses less CO2 😅
03.05.2025 18:58 — 👍 8 🔁 0 💬 1 📌 0
No. The other libraries have different names (e.g RCSB Mol*, PDBe Mol*). Mol* = mol* = molstar
03.05.2025 10:08 — 👍 0 🔁 0 💬 0 📌 0

Google Colab
It would be nice if molviewspec would support trajectories because you could easily set up your scene from python and then just use an iframe viewer. colab.research.google.com/drive/1O2Tld...
But seems only pdb/cif and map data so far: molstar.org/mol-view-spe...
02.05.2025 09:08 — 👍 1 🔁 0 💬 1 📌 0
Molstar XTC
Created on https://plnkr.co: Helping you build the web.
The problem is that there are various libraries packaging mol* into various viewers. I don't think the PDBe one supports xtc files, Mol* supports it though. Here is an example using native mol* taken from the quarto-molstar package.
plnkr.co/edit/TjTgopo...
02.05.2025 09:01 — 👍 1 🔁 0 💬 2 📌 0
If you want to customize this viewer it is much more complicated.
02.05.2025 06:59 — 👍 1 🔁 0 💬 1 📌 0
PDBe Molstar plugin - Basic - v3.2.0
Created on Plnkr: Helping developers build the web.
The various flavours of Mol* should be pretty easy to integrate but can be a bit more tricky if you use SSR in a modern webstack. Example: embed.plnkr.co/plunk/eMPsfu... You need to convert the trajectory into e.g a PDB trajectory. Try with a NMR structure like 3GB1.
02.05.2025 06:56 — 👍 4 🔁 0 💬 1 📌 0
Yes but it requires a certain level of understanding of copyright. So many papers that don't even acknowledge 3rd party material properly... I'm looking for funding to build a fully CC0 db to make this easier... currently we spend ~10-20 Mio $ annually on BioRender globally 😔
22.03.2025 21:36 — 👍 2 🔁 0 💬 0 📌 0
CC BY is great but also requires that we replace some of our tools. Many currently published CC BY papers are actually not fully CC BY because of BioRender www.chemistryworld.com/news/thousan...
19.03.2025 17:30 — 👍 4 🔁 2 💬 1 📌 0
Might be related to this: bsky.app/profile/raph...
02.03.2025 18:08 — 👍 1 🔁 0 💬 1 📌 0
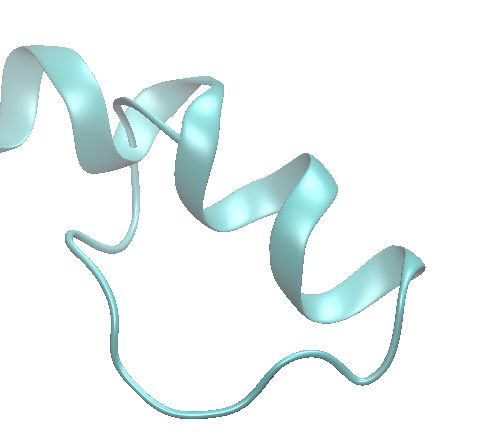
Installation and running it was a breeze 🤗 Diversity of the generated structures is quite good. Quality is a bit hit and miss. Most samples at default params have at least one or two weird C-N peptide bonds >1.6A/<1.1A and also had a few (<0.5%) samples with quite severe clashes.
21.02.2025 10:55 — 👍 1 🔁 0 💬 0 📌 0
Yes that's true but afaik ZeroGPU might get Docker support as well (although I think there is no timeline). You can also ask them for the old community GPU grant which works with generic docker images but has different set of limitations especially for high traffic applications.
17.02.2025 16:32 — 👍 0 🔁 0 💬 0 📌 0

Docker Spaces
We’re on a journey to advance and democratize artificial intelligence through open source and open science.
One can build quite complex interfaces with gr.Blocks() in my opinion. Are you using Blocks? Blocks works basically like flexbox layouts. HF spaces also supports generic Docker containers (just expose a port and you use anything you like huggingface.co/docs/hub/spa...)
17.02.2025 10:14 — 👍 1 🔁 0 💬 1 📌 0
In earlier work, we already analyzed how AllMetal3D given a structural input and no information about metal stoichiometry fares against co-folding methods given metal stoichiometry but no structure: doi.org/10.1101/2024...
10.02.2025 09:35 — 👍 0 🔁 0 💬 0 📌 0
We compared to a few other tools that perform either metal identity prediction or location prediction and found that is very important to include negative classes during training and also cluster metal ions by biological similarity.
10.02.2025 09:35 — 👍 0 🔁 0 💬 1 📌 0

ChimeraX preview
We've made available a ChimeraX extension and a Webapp to easily run predictions. AllMetal3D is now also easily installable using pip.
ChimeraX: cxtoolshed.rbvi.ucsf.edu/apps/chimera...
HuggingFace: huggingface.co/spaces/simon...
Docs for Python Package:
lcbc-epfl.github.io/allmetal3d/
10.02.2025 09:35 — 👍 0 🔁 0 💬 1 📌 0
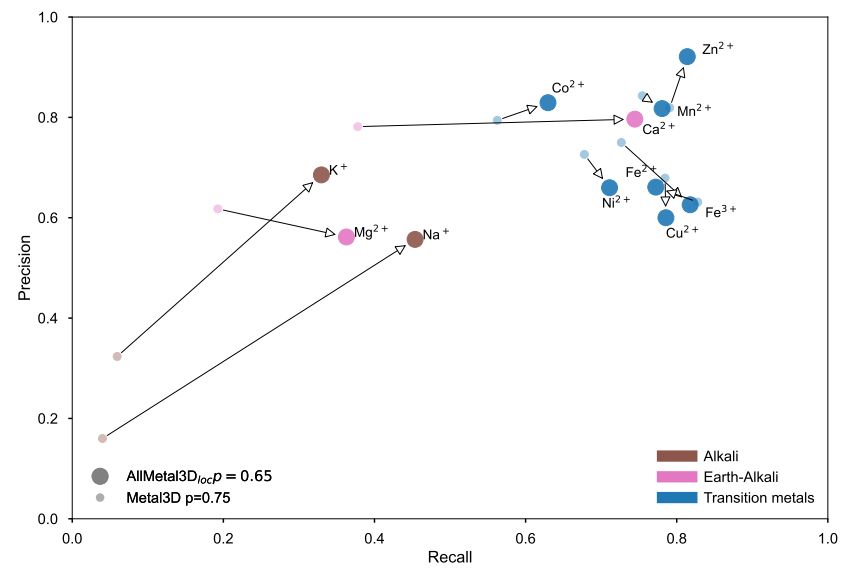
Performance of AllMetal3D vs. Metal3d.
Excited to share our new AllMetal3D model building on Metal3D. AllMetal3D predicts the location, identity and geometry of the major biologically relevant metal ions doi.org/10.1101/2025...
10.02.2025 09:35 — 👍 17 🔁 5 💬 1 📌 0

It’s been a tough few weeks. My 10yo daughter was diagnosed with a very rare, aggressive cancer called interdigitating dendritic cell sarcoma (IDCS). I’m reaching out to identify clinicians/patients who have encountered pediatric IDCS or other (non-LCH) dendritic or histiocytic sarcomas cases.
08.02.2025 21:21 — 👍 1017 🔁 863 💬 82 📌 32
Gifs are the least accessible form of interactive content. The best would be interactive HTML figures.
27.01.2025 07:24 — 👍 2 🔁 0 💬 1 📌 0
I've seen cases with open code for the webserver without the actual model or method used for the research published as well ... Code review should be part of peer review.
21.01.2025 05:46 — 👍 5 🔁 0 💬 0 📌 0
Nice tool. I use visidata which reads Json, CSV and other formats and even allows to do basic calculations.
05.01.2025 06:03 — 👍 1 🔁 0 💬 1 📌 0
Good for you, the people who published these articles did as well. Nevertheless, the company says the people who paid them for a license violated their copyright and need to correct their article when I asked for reusing it under the stated license. Is that open science for you?
22.12.2024 19:26 — 👍 1 🔁 0 💬 0 📌 0
I asked BioRender about reusing illustrations from a CC BY article some time ago and they told me the license of the article is not correct and a correction will have to be issued. Do you want to track down all authors to correct all 153 articles in PloS Biology that make use of BioRender?
22.12.2024 17:04 — 👍 1 🔁 0 💬 1 📌 0
Bioicons - high quality science illustrations
Bioicons contains free science illustrations for biologists, chemists, machine learning under permissive CC0, CC BY or MIT License.
Totally, I have been building bioicons.com since a few years. However, more funding for truly open science illustrations will not happen if stakeholders like journals continue to believe BioRender's ridiculous attempts to salvage their business model.
22.12.2024 17:04 — 👍 1 🔁 0 💬 1 📌 0
Why does PLOS enforce CC BY for all 3rd party content if not to allow reuse in other journals without filling permission forms or hidden copyright complexities?
22.12.2024 16:59 — 👍 1 🔁 0 💬 0 📌 0
Also BioRender's new "CC BY" license requires that users put a specific link where above conditions are displayed because they have to explain that they differ from standard CC BY. (CC BY doesn't allow modification of the conditions). This is not the case for this PLOS article.
22.12.2024 16:59 — 👍 0 🔁 0 💬 1 📌 0
In the fictional universe of the BioRender lawyers they can claim they are compatible with CC BY but fact is the figures created in BioRender cannot be reused under CC BY because they say one needs a paid account which is against the CC BY terms (compare BioRender vs. @creativecommons.bsky.social).
22.12.2024 16:59 — 👍 0 🔁 0 💬 1 📌 0
Frontier models for all molecules of life.
Omics & Software Eng 🧬👨🏽💻 | UC-Berkeley 🐻👨🏽🔬 | Current LSRF HHMI Awardee🎓 | HHMI Gilliam Alumn🧑🎓
https://linktr.ee/jlsteenwyk 🔗🌲
jlsteenwyk.com 🔗📍
Associate Researcher in Computational Life Sciences at FHNW. I'm interested in Molecular Simulation, Machine Learning and Bioinformatics to find out a thing or two about RNA, Alternative Splicing in Cancer and more :)
Lab head at the University of Melbourne. Studying the structure and biochemistry of microbial membrane proteins and metalloenzymes.
BWF CASI Fellow @Columbia | 2022 Jane Coffin Childs Fellow | PhD @UCBerkeley | BS @TuftsUniversity | Systems biophysics via integrative ML- and physics-based models
Assistant Professor in Biochemistry & Molecular Pharmacology, Baylor College of Medicine. CryoEM, GPCRs, Membrane Proteins, Comp Chem, SBDD.
Operated by @shervinnia.bsky.social and inspired by the now-defunct bot by Frédéric Poitevin and @gati.bsky.social. Suggestions welcome.
Weekly #RobSelects
Assistant Professor for Computer-Aided Organic Synthesis at the Stratingh Institute for Chemistry, University of Groningen
(he/him/his)
[bridged from https://mstdn.science/@robpollice on the fediverse by https://fed.brid.gy/ ]
My name is Jorge Camões, I'm a #dataviz consultant, trainer, and author of the book Data at Work amzn.to/2i7Z0KO. #Excel, #PowerBI & #PowerQuery, #Tableau. Based in Lisbon, Portugal. wisevis.com
Chief Digital Officer @PLOS.org. Previously responsible for Wellcome Collection’s digital transformation, Director of Product at Springer Nature, BBC /nature, /programmes /music &c. Oenophile and Type 1 Diabetic.
https://tomscott.name
AITHYRA is a new dynamic research institute for biomedical AI in Vienna. AITHYRA seeks to build Europe’s premier institute for AI-driven biological and medical research, uniting computer scientists, engineers, and biologists in a collaborative environment.
Independent AI researcher, creator of datasette.io and llm.datasette.io, building open source tools for data journalism, writing about a lot of stuff at https://simonwillison.net/
Chemical biologist @ Max Planck Institut for Medical Research in Heidelberg
Biophysics; protein dynamics/folding using smFRET and HDX-MS.
https://jhsmit.org/
PhD student@MIT
https://sites.google.com/view/yehlincho/home
Computational biologist; Associate Prof. at University of Wisconsin-Madison; Jeanne M. Rowe Chair at Morgridge Institute
Professor & Director leading @salibalab.bsky.social @unistuttgart.bsky.social & @fz-juelich.de. Scientist on sustainable energy. Prior: @tuda.bsky.social @unifr.bsky.social #EPFL @ox.ac.uk.
Editor-in-Chief of EES Solar @ees-journals.rsc.org.








