Thanks! Happy to hear any feedback 🙂
21.10.2025 15:03 — 👍 0 🔁 0 💬 0 📌 0
Bottom line: dLEM shows we don't have to choose between mechanistic insight and predictive power.
By reformulating loop extrusion as a differentiable process, we get:
✅ Interpretability
✅ Scalability
✅ Perturbation prediction
✅ Integration with ML
10/10
21.10.2025 14:04 — 👍 0 🔁 0 💬 1 📌 0
The parameter reduction compared to other deep learning models in deep dLEM is wild 🤯:
- 280× fewer than C.Origami
- 750× fewer than Orca
Yet we achieve competitive accuracy!
Why? Because we hard-coded the biophysics instead of making the model learn it.
9/10
21.10.2025 14:03 — 👍 0 🔁 0 💬 1 📌 0
We took dLEM to the next level: deep dLEM 🧠
We embedded dLEM as a biophysical layer in a neural network that learns from:
- DNA sequence
- Chromatin accessibility (DNase/ATAC)
Result: Sequence → structure prediction that stays mechanistically grounded
8/10
21.10.2025 14:01 — 👍 0 🔁 0 💬 1 📌 0
Example 2: CTCF depletion
→ We systematically reduced CTCF coefficients in our linear model
→ Optimal prediction at α=0.33, matching the 33% of CTCF peaks that remain after auxin treatment
The model reveals residual CTCF is still functionally active!
7/10
21.10.2025 14:01 — 👍 0 🔁 0 💬 1 📌 0
Because dLEM parameters are physically interpretable, we can do something deep learning can't: predict perturbation effects!
Example: WAPL depletion
→ We modified just the detachment rate
→ Successfully predicted emergence of new loops & extended stripes
21.10.2025 14:00 — 👍 0 🔁 0 💬 1 📌 0
What do we learn from these velocity profiles?
😌 CTCF shows expected directional asymmetry (blocks L or R depending on orientation)
😲 BUT: Transcription machinery (H3K4me3, H3K36me3, H3K4me1,...) ALSO modulates cohesin dynamics!
CTCF isn't the whole story 👀
5/10
21.10.2025 13:58 — 👍 0 🔁 0 💬 1 📌 0
dLEM acts as an encoder-decoder.
📥 ENCODER: Compress complex 2D Hi-C maps into simple 1D velocity profiles
📤 DECODER: Reconstruct contact maps from these profiles
It's dimensionality reduction, but with built-in biophysics!
4/10
21.10.2025 13:56 — 👍 0 🔁 0 💬 1 📌 0
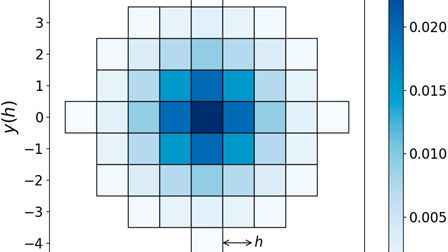
💡The key insight: While contact maps are 2D, loop extrusion is inherently 1D—cohesin walks along chromatin!
dLEM captures this with two velocity profiles:
→ L: leftward cohesin speed
→ R: rightward cohesin speed
Obstacles (like CTCF) = dips in velocity 📉
3/10
21.10.2025 13:56 — 👍 0 🔁 0 💬 1 📌 0
The challenge: Current methods face an impossible tradeoff.
❌ Polymer simulations = mechanistic, but not scalable
❌ Deep learning = predictive, but black-box
❌ Summary metrics = compressed, but mechanism-agnostic
We needed something that's mechanistic, interpretable, AND scalable.
✅ Enter dLEM!
2/10
21.10.2025 13:54 — 👍 0 🔁 0 💬 1 📌 0
✨New preprint!✨
We built dLEM - a differentiable Loop Extrusion Model that bridges biophysics and machine learning for 3D genome folding.
dLEM makes loop extrusion learnable and interpretable—predicting how genomes fold and how they respond to perturbations.
🧵
21.10.2025 13:51 — 👍 4 🔁 3 💬 2 📌 0
8/8 I had so much fun working on this paper with Ivo @mosaicgroup.bsky.social group at @mpicbg.bsky.social! Thanks for all the support, patience, and freedom to explore! # CompBio #Biophysics #PhDLife #Science #Research
20.12.2024 13:05 — 👍 1 🔁 0 💬 1 📌 0
7/8 There's more to explore, especially for 2D systems (like membranes), but this changes how we use these models to study cellular processes!
20.12.2024 13:04 — 👍 1 🔁 0 💬 1 📌 0
6/8 When these models "lose" reactions, they're actually capturing a real physical transition to the diffusion-limited regime. This opens new possibilities for studying complex molecular systems.
20.12.2024 13:03 — 👍 0 🔁 0 💬 1 📌 0
5/8 On top of that, we find that different diffusion models (like GRDME vs traditional RDME) correspond to different molecular sizes on the same grid. So even with this interpretation of RDME, we can model systems with molecules of different sizes!
20.12.2024 13:03 — 👍 0 🔁 0 💬 1 📌 0
4/8 Our key finding is that grid size sets molecular size! Smaller boxes —> smaller molecules —> need to get closer to react—> fewer reactions. This perfectly matches the theory for diffusion-limited systems.
20.12.2024 13:02 — 👍 0 🔁 0 💬 1 📌 0
3/8 But what if the grid size itself represents the reaction radius - the distance at which molecules can react? Then fewer reactions when boxes are smaller would be exactly what should happen physically! 💡
20.12.2024 13:01 — 👍 0 🔁 0 💬 1 📌 0
2/8 In grid-based models (RDME/GRDME), molecules jump between boxes and react when in the same box. As we make boxes smaller, reactions become less frequent - for years, this was seen as a flaw! 🤔
20.12.2024 13:01 — 👍 0 🔁 0 💬 1 📌 0
7/7 This work led to some surprising insights about how these models actually represent physical reality, particularly the 'reaction loss' phenomenon... and that's a story of the just published paper! 😉 #CompBio #Biophysics #PhDLife #Science #Research
19.12.2024 21:18 — 👍 0 🔁 0 💬 0 📌 0
6/7 Here's where it gets interesting: RDME "loses" bimolecular reactions at very fine grid resolutions. Well, so does GRDME! We derived a rule of thumb to predict this limit - turns out GRDME's limit is higher by the same factor as its speedup.
19.12.2024 21:18 — 👍 0 🔁 0 💬 1 📌 0
5/7 Both RDME and GRDME show second-order convergence of the diffusion error with grid resolution. While GRDME has a slightly larger error, it runs up to 6x faster in 3D (or even more if you increase the smoothing length)! There's no free lunch in computing, but this is a pretty good deal!
19.12.2024 21:18 — 👍 0 🔁 0 💬 1 📌 0
4/7 PSE naturally comes with a 'smoothing length' parameter (the spread of the Gaussian). By repurposing it for GRDME, we get a built-in way to tune the trade-off between speed and accuracy!
19.12.2024 21:17 — 👍 0 🔁 0 💬 1 📌 0
3/7 Our new method, GRDME, lets molecules make "Gaussian jumps" - like confetti 🎊, they can land in boxes further away, with closer boxes being more likely! The math comes from Particle Strength Exchange (PSE), a discretization technique from the particle methods community.
19.12.2024 21:17 — 👍 0 🔁 0 💬 1 📌 0
2/7 I worked with RDME (Reaction-Diffusion Master Equation) - a method that divides space into a grid of boxes and tracks molecules moving between them. Molecules can only jump to adjacent boxes —> it approximates diffusion with finite differences - simple and accurate, but oh so slow! 🐌
19.12.2024 21:16 — 👍 0 🔁 0 💬 1 📌 0
My second PhD paper just got published! 🎉 So, it’s good time to introduce my PhD work to Blue Sky.
19.12.2024 21:13 — 👍 0 🔁 0 💬 0 📌 0
Great to be here! I guess I should introduce myself: I’m a postdoc (Chikina lab), using physical modeling and machine learning to study transcription and genome organization. But, I’d really like to get into plant science and tackling climate change!
14.11.2024 15:09 — 👍 3 🔁 0 💬 0 📌 0
disaster lesbian on the apocalypse beat @wired.com . very tall with a very short dog. they/them. pitch me: molly_taft@wired.com, signal: @mollytaft.76. mollytaft.com
The Marine Biological Laboratory (MBL), founded in 1888, is an international center for research & education in biology & ecology in Woods Hole, MA. An affiliate of the University of Chicago. www.mbl.edu
Lego fan for as long as I can remember...
I'll be posting any mocs i build here
Lego is a big passion of mine along side art so if your interested in the wonderful world of lego please stick around! https://linktr.ee/Zanzart
We aim to serve society by making science open, inclusive, and accessible. Also @500womensci on Instagram and Facebook. Find out more: https://500womenscientists.org/
https://linktr.ee/500womenscientists
Pittsburgh and Western Pennsylvania's leading source for news and analysis. Support local journalism by subscribing at http://my.post-gazette.com/activate
View our starter pack: https://go.bsky.app/Gaxi8Zc
TribLIVE.com covers Pittsburgh and Western Pennsylvania news, sports, politics, entertainment and more.
PhD student @mosaicgroup.bsky.social working on machine learning for understanding the spatial organization of living systems from images.
MPI-CBG, Paul Langerhans Institute (PLID) -Developmental and stem cell biologist- Physics of Life
Mathematicians developing mathematics and methods to look at living systems differently.
Colocated at @mpi-cbg.de and @csbdresden.bsky.social in Dresden, Germany.
proud papa | group leader team stembryo @ MPI-CBG Dresden | re- and deconstructing (st)embryos | #stembryogenesis | solves problems in lab & climbing hall | views mine
CMU-Pitt Computational Biology PhD Candidate
Cartoonist and creator of The Oatmeal and Exploding Kittens https://linktr.ee/theoatmeal
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Assistant Professor UMass Chan, Board of Directors NumFOCUS
Previously IMP Vienna, Stanford Genetics, UW CSE.
Assistant Professor, Biochemistry Department, Transcription & Chromatin biology, Outdoor enthusiast, Dog lover (otters too) 🇵🇭🇺🇲🇨🇦
Professor at the University of British Columbia @sbmeubc.bsky.social
Specially Appointed Professor at Osaka University PRIMe
https://yachie-lab.org
Assistant Professor, UBC school of Biomedical Engineering. Trying to enable personalized medicine by solving gene regulatory code.
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
group leader @ HHMIJanelia, #neuroscience + AI 🔬 #cellpose | diversity and open-science for better science | Ⓥ | she/her | https://mouseland.github.io
Biophotonics, Biophysics & Innovation
@KITkarlsruhe