
Like regulatory genomics? Don’t miss this very fun meeting! May 7 is the deadline for early registration and abstract submission www.asbmb.org/meetings-eve...
07.05.2025 01:57 — 👍 15 🔁 5 💬 0 📌 2@mlweilert.bsky.social
bioinformatics, cats, deep learning, genomics, Zeitlinger Lab ((all views are mine alone))

Like regulatory genomics? Don’t miss this very fun meeting! May 7 is the deadline for early registration and abstract submission www.asbmb.org/meetings-eve...
07.05.2025 01:57 — 👍 15 🔁 5 💬 0 📌 2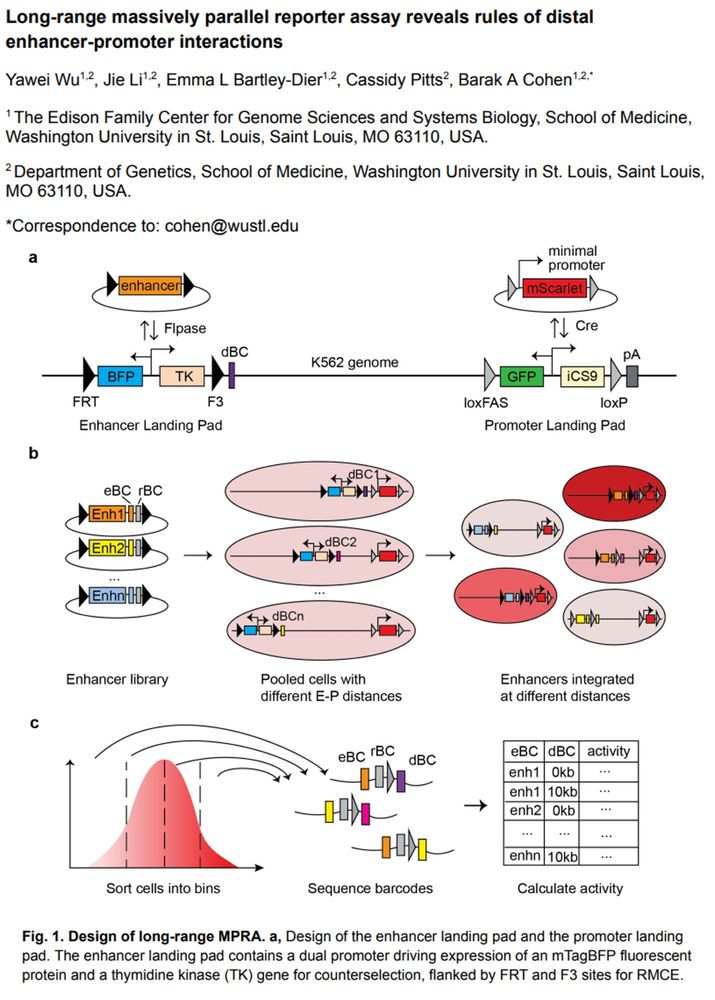
From the BioRxiv pre-print article "Long-range massively parallel reporter assay reveals rules of distal enhancer-promoter interactions" by Yawei Wu et al. Fig. 1. Design of long-range MPRA. a, Design of the enhancer landing pad and the promoter landing pad. The enhancer landing pad contains a dual promoter driving expression of an mTagBFP fluorescent protein and a thymidine kinase (TK) gene for counterselection, flanked by FRT and F3 sites for RMCE. available under aCC-BY-NC 4.0 International license. was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made bioRxiv preprint doi: https://doi.org/10.1101/2025.04.21.649048; this version posted April 22, 2025. The copyright holder for this preprint (which The promoter landing pad expresses the mEmerald fluorescent protein and iCaspase9 for counterselection and is flanked by loxFAS and loxP sites for orthogonal RMCE. b-c, Workflow of long-range MPRA. eBC: enhancer barcode; rBC: random barcode; dBC: distance barcode. A barcoded enhancer library is integrated into pooled long-range landing pad cells through Flpase-facilitated recombination. After counterselection with Ganciclovir, cells with successful integration are enriched. Cells are sorted into different bins based on mScarlet fluorescence level. Genomic DNA is extracted from each bin; barcodes are amplified and sequenced. Reporter gene expression of each enhancer at each distance is calculated as the weighted average of fluorescence levels across bins.
🚨PRE-PRINT 🧪🧬🖥️👩🔬
Long-range massively parallel reporter assay reveals rules of distal enhancer-promoter interactions
From Barak Cohen's lab at @washu.bsky.social
Read the pre-print 👇
doi.org/10.1101/2025...
Learn more about the research from the Cohen lab: bclab.wustl.edu
Our new preprint is out! Want to better visualize what your sequence-to-function profile learned? Here is PISA. It also comes in a new BPNet package, which can be used to train many genomics data sets, including MNase-seq data.
08.04.2025 13:31 — 👍 26 🔁 8 💬 1 📌 0
Very proud of two new preprints from the lab:
1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...

We're thrilled that Investigator @juliazeitlinger.bsky.social is co-chairing the 2025 @asbmb.bsky.social meeting in #KansasCity, which will delve into the core processes of #geneexpression from #developmental and evolutionary perspectives.
Read more: bit.ly/41Czvqy
This a great interdisciplinary gene expression meeting, where everyone is welcome. Please consider attending and submitting your abstract: www.asbmb.org/meetings-eve...
04.03.2025 14:10 — 👍 12 🔁 6 💬 0 📌 1dear god
23.02.2025 14:44 — 👍 0 🔁 0 💬 0 📌 0Or just rename every one to start with B. Bavocado, Bragonfruit, Bomegranate 🤭
19.02.2025 01:53 — 👍 1 🔁 0 💬 1 📌 0
Thermodynamic principles link in vitro transcription factor affinities to singlemolecule chromatin states in cells
www.biorxiv.org/content/10.1...
5 minutes on this platform and it's already clear leaving Twitter was a great call, glad to see everyone <3
29.01.2025 00:35 — 👍 4 🔁 0 💬 1 📌 0