
How did eukaryotic cells with complex architecture evolve from simpler prokaryotic cells? DNA analyses offer possible answers
go.nature.com/4sEMwLH
@derdunk.bsky.social
Studying the evolution of complexity through modelling and phylogenomics

How did eukaryotic cells with complex architecture evolve from simpler prokaryotic cells? DNA analyses offer possible answers
go.nature.com/4sEMwLH
Less than two 2 weeks to apply for this PhD position in my group.
Please do not miss the opportunity to apply if the topic is of interest to you 👇

@guyleonard.bsky.social et al. report a near-complete genome and transcriptome sequence dataset for green algae host Paramecium bursaria, an endosymbiotic model system.
🔗 doi.org/10.1093/gbe/evaf183
#genome #evolution
Very nice documentary about the origins of complex life with my PhD advisor Berend Snel @binfutrecht.bsky.social!
09.10.2025 08:46 — 👍 12 🔁 7 💬 0 📌 0LMB PhD studentship available in the Baum lab to use informatics to infer cell structure from genomic data in collaboration with the brilliant Tom Williams. If you write to me and I don’t reply - try again - the spam filter is hungry…
02.10.2025 09:04 — 👍 17 🔁 19 💬 1 📌 2
Photo showing the inside atrium of the new Life and Mind Building
Oxford Biology is growing 📢
We’re appointing 3 Associate Professors in:
🌱 Plant Sciences
🦉 Animal Behaviour
🔬 Molecular Cell Biology
3 fields. 3 opportunities. One new home for Oxford Biology.
Learn more 👉 bit.ly/41S2Tc7
Apply now 👉 bit.ly/488CNW3

Protein structures are not inherently robust, but natural proteins are substantially more robust than random sequences, representing a clear signature of adaptation. Mutational robustness is relatively low in newborn and viral proteins, and provides a new perspective on protein evolution.
01.09.2025 10:35 — 👍 0 🔁 0 💬 0 📌 0
RNA structures are inherently evolvable and robust to mutations: random RNA molecules already show the same level of robustness and a similar distribution of secondary structures as natural RNA molecules. Hence, it is difficult to detect signatures of selection on the distribution of RNA structures.
01.09.2025 10:35 — 👍 0 🔁 0 💬 1 📌 0Excited with two new preprints on evolution of molecular structure, in RNA (doi.org/10.1101/2025..., @n-martin.bsky.social) and proteins (doi.org/10.1101/2025...). Both RNA and protein structures are often conserved across evolution, but the underlying reasons are very different.
01.09.2025 10:35 — 👍 8 🔁 3 💬 1 📌 0And thanks to all poster presenters, including Moisès Bernabeu whose poster was in the top 5 of best posters at #ESEB2025. Congrats!
22.08.2025 14:27 — 👍 7 🔁 3 💬 0 📌 0Sam von der Dunk: we know that RNA structure is robust to mutation yet evolvable. What about protein structure? Take advantage of ESMfold https://doi.org/10.1126/science.ade2574
Natural proteins are robust to mutations, whereas random amino-acid sequences are not. Even disordered natural […]




Great to see so many people at our symposium om the origin of eukaryotes. Thanks to all the speakers! #ESEB25 @julianvosseberg.bsky.social
21.08.2025 16:23 — 👍 12 🔁 2 💬 0 📌 1
Interactive symbiosis in silico (back) and in vivo (front). #ESEB25
19.08.2025 22:25 — 👍 20 🔁 3 💬 0 📌 0We have found before, including in different models, that evolutionary conflict leads to better adaptive solutions. These challenges best expose the creativity of evolutionary processes. Or, borrowing from Nietzsche: "What doesn't kill you makes you stronger" (at the population level)!
05.08.2025 09:35 — 👍 1 🔁 0 💬 0 📌 0Our main finding: Mixing of endosymbionts (sex) is dangerous but can stimulate beneficial regulatory integration of host and symbiont!
05.08.2025 09:35 — 👍 1 🔁 0 💬 1 📌 0
The exciting project of Alkmini Zania on the impact of sex on endosymbiosis reveals complex evolutionary dynamics. What could this mean for early eukaryotes? www.biorxiv.org/content/10.1...
05.08.2025 09:35 — 👍 5 🔁 0 💬 1 📌 0
🚨 Join us at TBB Utrecht by applying for this evo-devo PhD position! 🚨
www.uu.nl/en/organisat...
One week left to submit your abstract! @eseb2025.bsky.social
17.04.2025 10:16 — 👍 7 🔁 3 💬 0 📌 0
Share your work on host-associated microbiomes in our symposium @eseb2025.bsky.social - with speakers Hassan Salem @hassansalem.bsky.social and Carola Petersen. Abstract deadline April 25th. Florent, Klara, and I would love to see you there!
13.04.2025 18:19 — 👍 19 🔁 6 💬 1 📌 5
Join us in at #ESEB2025 Barcelona for our symposium on microbial adaptation to changing conditions. Organised by @mtoll8.bsky.social @javierdelafuente.bsky.social and me. Amazing invited speakers @saramitri.bsky.social and Itzik Mizrahi!! @eseb2025.bsky.social
13.03.2025 17:30 — 👍 47 🔁 24 💬 2 📌 0
#ESEB2025 Exciting symposium alert: (S50) Unravelling the origin of eukaryotes. 🦑🌴 With speakers Purificación López-García and Mark Field. We are looking forward to your abstract submissions! eseb2025.com/list-of-symp... @julianvosseberg.bsky.social
17.03.2025 18:09 — 👍 8 🔁 10 💬 0 📌 1In our study, we found that genome organization emerges, in which the positions of regulatory genes along the chromosome correlate with their temporal activity during the cell cycle...
17.03.2025 12:44 — 👍 0 🔁 0 💬 0 📌 0
Here's a cool article doi.org/10.1038/s415..., which supports my evolutionary model of a prokaryotic cell cycle (doi.org/10.1093/gbe/...), in which we allow evolution to exploit the changes in gene dosage induced by replication to improve cell-cycle regulation.
17.03.2025 12:44 — 👍 1 🔁 0 💬 1 📌 0
I would really appreciate a wide sharing of this post, as the candidate profile I am looking for is outside of my regular circle. Thank you so much. More info and how to apply here: uu.varbi.com/se/what:job/... (2/2)
10.12.2024 07:42 — 👍 3 🔁 15 💬 1 📌 0New MRC-funded postdoc with me on comparative bacterial genomics/phylodynamics @roslininstitute.bsky.social university of Edinburgh. Great multidisciplinary team and collaboration with partners at Shanghai Jao Tong University.
elxw.fa.em3.oraclecloud.com/hcmUI/Candid...
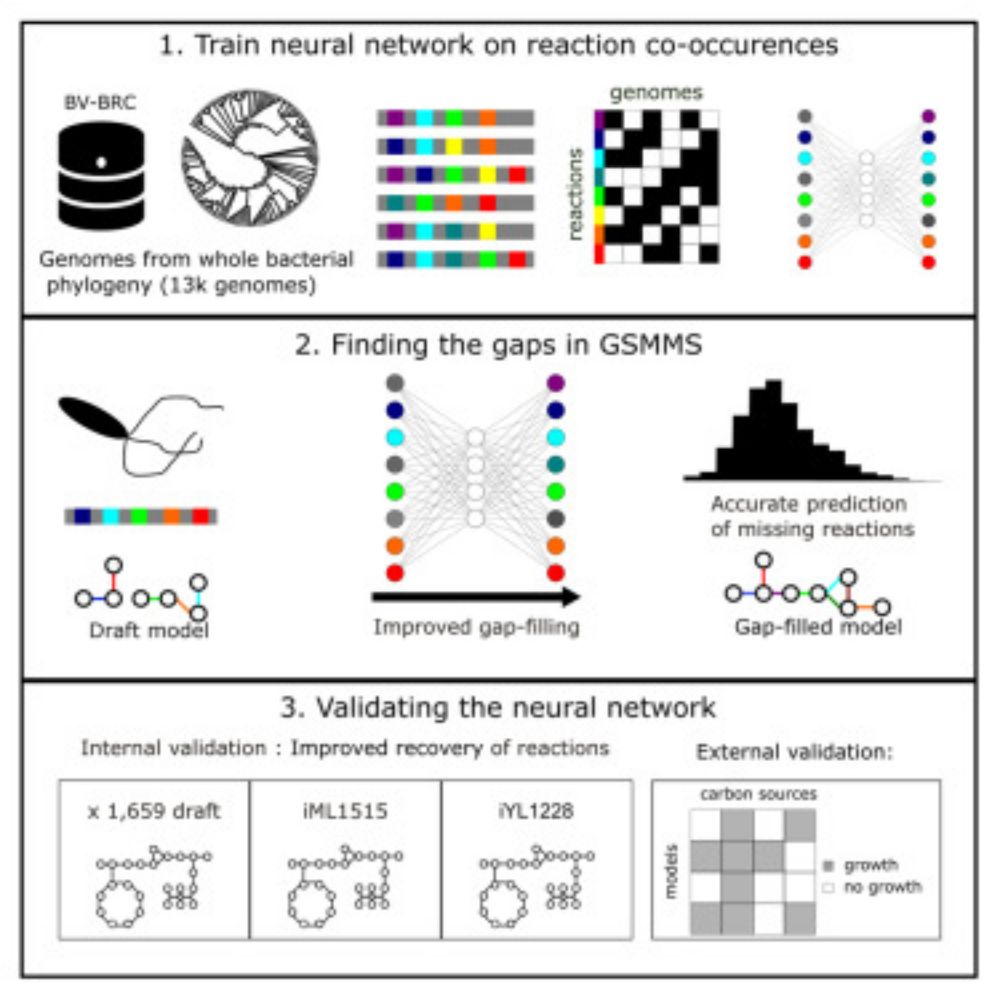
Exciting news! 🎉 My article is out in iScience presenting DNNGIOR, a deep learning tool advancing gap-filling in metabolic models. Huge thanks to co-authors @cmelkonian.bsky.social, @hariszaf.bsky.social, Daniel Garza, Andreas Haas and @bedutilh.bsky.social. Read at shorturl.at/REOm8
26.11.2024 12:25 — 👍 10 🔁 4 💬 0 📌 1Hope that helps?
26.02.2024 12:08 — 👍 0 🔁 0 💬 0 📌 0Although we enforce the endosymbiosis, host and symbiont normally adapt in such a way that preserves their regulatory autonomy. Only when we include regulatory conflicts, do host and symbionts evolve to communicate and to establish control mechanisms between them.
26.02.2024 12:08 — 👍 0 🔁 0 💬 1 📌 0Regulatory conflicts between host and symbionts (i.e. signaling molecules of host and symbiont interfering with each other) promote host--symbiont integration, providing a narrative for how a transition in individuality may take place in a tight endosymbiotic relationship.
26.02.2024 12:05 — 👍 0 🔁 0 💬 1 📌 0... Surprisingly, these regulatory conflicts inspire evolution to come up with new solutions for host--symbiont cell-cycle coordination. Host and symbiont evolve communication and various control strategies, and in some cases even manage to synchronize their cell cycles.
26.02.2024 11:58 — 👍 0 🔁 0 💬 0 📌 0