Overall, ARG inference tools have demonstrated remarkable robustness to computational phasing errors. While various practical challenges may limit ARG inference quality, we believe that computational phasing inaccuracies should not be a problem of concern.
7/7
26.11.2025 17:08 — 👍 2 🔁 0 💬 0 📌 0

Under a selective sweep model, we saw little difference in branch-length-based diversity between ARGs inferred from true and computationally phased haplotypes, demonstrating the practicality of applying ARGs inferred from unphased data in evolutionary analyses.
6/7
26.11.2025 17:08 — 👍 0 🔁 0 💬 1 📌 0

Under the CEU bottleneck model, we observed consistently minor effects of phasing errors. The difference between ARG inference methods is much larger than the difference in performance between using true haplotypes and computationally phased haplotypes.
5/7
26.11.2025 17:08 — 👍 0 🔁 0 💬 1 📌 0

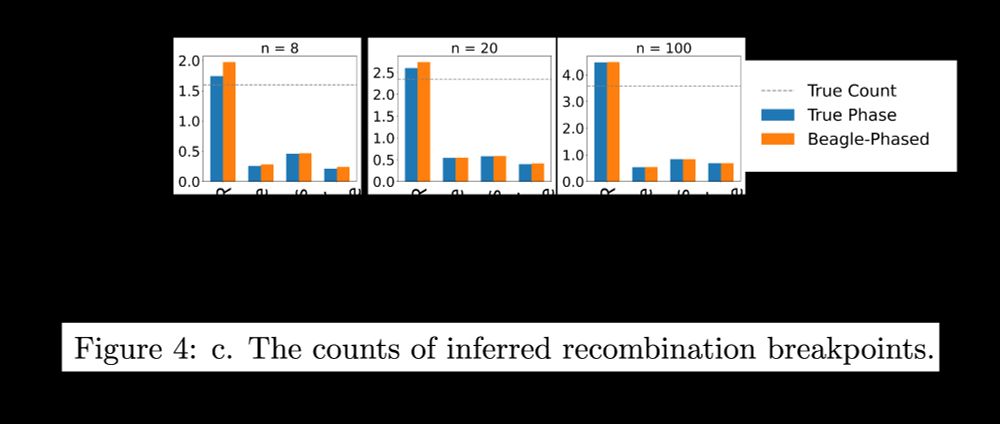
Here, we show that ARG inference remains robust to phasing errors even at incredibly small sample sizes (8 haplotypes). We simulated ARGs and VCFs under various demographic models, simulated phasing errors, and compared ARGs inferred from true and computationally phased haplotypes.
3/7
26.11.2025 17:08 — 👍 2 🔁 0 💬 1 📌 0

ARGs are powerful tools in population genetics. However, ARG inference generally requires phased haplotypes. Phasing quality is limited at small sample sizes, making it difficult in non-model organisms due to the usually limited sample size and a lack of reference panels.
2/7
26.11.2025 17:08 — 👍 2 🔁 0 💬 1 📌 0
Overall, ARG inference tools have demonstrated remarkable robustness to computational phasing errors. While various practical challenges may limit ARG inference quality, we believe that computational phasing inaccuracies should not be a problem of concern.
7/7
26.11.2025 16:47 — 👍 0 🔁 0 💬 0 📌 0

Under a selective sweep model, we saw little difference in branch-length-based diversity between ARGs inferred from true and computationally phased haplotypes, demonstrating the practicality of applying ARGs inferred from unphased data in evolutionary analyses.
6/7
26.11.2025 16:47 — 👍 0 🔁 0 💬 1 📌 0

Under the CEU bottleneck model, we observed consistently minor effects of phasing errors. The difference between ARG inference methods is much larger than the difference in performance between using true haplotypes and computationally phased haplotypes.
5/7
26.11.2025 16:47 — 👍 0 🔁 0 💬 1 📌 0

Here, we show that ARG inference remains robust to phasing errors even at incredibly small sample sizes (8 haplotypes). We simulated ARGs and VCFs under various demographic models, simulated phasing errors, and compared ARGs inferred from true and computationally phased haplotypes.
3/7
26.11.2025 16:47 — 👍 0 🔁 0 💬 1 📌 0

ARGs are powerful tools in population genetics. However, ARG inference generally requires phased haplotypes. Phasing quality is limited at small sample sizes, making it difficult in non-model organisms due to the usually limited sample size and a lack of reference panels.
2/7
26.11.2025 16:47 — 👍 1 🔁 0 💬 1 📌 0
Population and evolutionary genetics @UCDavis. Posts, grammar, & spelling are my views only. He/him. #OA popgen book https://github.com/cooplab/popgen-notes/releases
Evolutionary Biologist at Stanford. Rapid Evolution, Adaptation, and Genomics. Open Science advocate.
Bióloga do IB-USP 07 🇧🇷 PhD UCBerkeley 🇺🇸 now postdoc at GEE-UCL 🇬🇧
Evolutionary genomics 🧬 🌳 popgen, ARGs, balancing selection. Interested in teaching, conservation. https://profiles.ucl.ac.uk/92078-debora-yoshihara-caldeira-brandt/
Associate professor at the Hebrew University of Jerusalem.
Statistical, population, and medical genetics; preimplantation genetic testing. Views my own.
http://scarmilab.org
https://jiawei-xing.github.io
Computational postdoc fellow at Siepel Lab, Cold Spring Harbor Lab. Interested in modeling transcriptional regulation by machine learning methods.
https://xzengcombio.github.io/
Postdoc at Siepel Lab, CSHL
Statistical genetics, phylogenetics, and optimization
PhD candidate at Siepel Lab, CSHL
Postdoc in Pritchard lab at Stanford University. Previous PhD co-advised by Yun Song and Rasmus Nielsen at UC Berkeley.
Undergraduate student studying cognitive neuroscience & biology @Washington University in St. Louis