
FLAMES is freely available from GitHub:
github.com/Marijn-Schip...
Enhanced PDF available here: rdcu.be/d9iQP
@mjschipper.bsky.social
Geneticist, Programmer and Science Enthousiast

FLAMES is freely available from GitHub:
github.com/Marijn-Schip...
Enhanced PDF available here: rdcu.be/d9iQP

We use FLAMES to prioritize 180 schizophrenia risk genes. We find that these genes are highly enriched in synaptic functions.
Clustering these genes based on relative expression throughout the lifetime shows that about one third of these genes are expressed strongest prenatally.

We benchmark our method against multiple tools, in different datasets (2 largest in fig). We find that FLAMES consistently outperforms other current gene prioritization methods.
Expert-curated = subset from: pubmed.ncbi.nlm.nih.gov/34711957/
ExWAS implicated from: pubmed.ncbi.nlm.nih.gov/37009933/
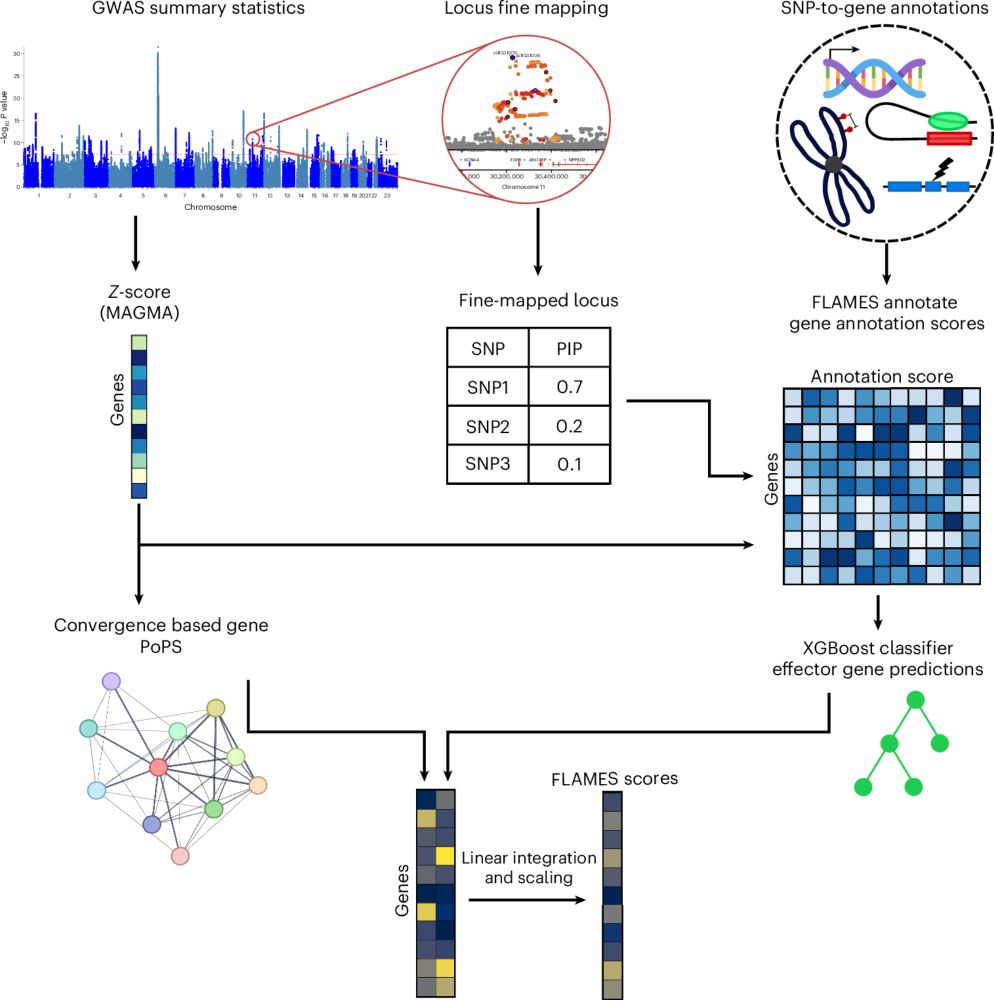
We trained an XGBoost classifier to predict the ExWAS gene in these loci based only on the SNP-to-gene annotations. Effectively asking the classifier what a causal gene looks like based on functional evidence.
We then reweight the XGBoost predictions with convergence-based evidence from PoPS.

FLAMES annotates 95% credible sets from fine-mapped GWAS loci with functional data linking SNPs to genes from over 20 sources.
We did this with 1181 loci which contain a gene also implicated by rare pLoF variants. We find these pLoF ExWAS genes enriched in functional annotations from GWAS SNPs.
Current integrative prioritization methods use either functional data (L2G, cS2G) or network convergence of GWAS signal (PoPS).
FLAMES combines both, by combining XGBoost predictions using functional evidence in the GWAS locus with PoPS predictions.
Incredibly proud to see our latest work out in Nature Genetics: www.nature.com/articles/s41...
Here we share our FLAMES framework, which predicts the effector genes in GWAS loci with state-of-the-art precision🔥
Special thanks to @daniposthu.bsky.social
A full thread describing findings below!