With the release of Boltz 2, is there already a review comparing the efficiency of different (new) docking tools? (I’ve already tried Pocket Vina, which is actually quite good for high-throughput)
31.08.2025 12:08 — 👍 3 🔁 1 💬 0 📌 1
A direct comparison with Boltz-2 hasn’t been done yet, but it would be interesting to see one between co-folding and the classical/hybrid docking benchmarks!
15.09.2025 21:15 — 👍 0 🔁 0 💬 0 📌 0

Illustration with overlaid text: The image shows a cancer cell in the bloodstream. The overlaid text appears in white, all-caps sans-serif font inside a dark blue rectangular box at the top left. It reads: “USING DEEP LEARNING FOR PRECISION CANCER THERAPY.” A small credit at the bottom right reads: “© Annie Cavanagh / Wellcome Collection.”
Nearly 50 new cancer drugs are approved each year – but which one fits which patient?
At the #mdcBerlin, @al2na.bsky.social’s team built Flexynesis, a deep learning toolkit to guide precision cancer care.
Learn more:
👉 www.mdc-berlin.de/news/press/u... 👈
12.09.2025 11:00 — 👍 9 🔁 4 💬 0 📌 0
is this how small molecules bind?? 😼
13.07.2025 03:40 — 👍 5 🔁 1 💬 1 📌 0
🚀 GPU-accelerated docking to P2Rank-predicted pockets
27.06.2025 07:25 — 👍 5 🔁 4 💬 0 📌 0

We benchmarked PocketVina across four widely used datasets (PDBbind, PoseBusters, Astex, DockGen), and introduce TargetDock-AI — a large-scale benchmark of >500K protein–ligand pairs with activity labels from PubChem.
(5/n)
26.06.2025 11:11 — 👍 2 🔁 1 💬 1 📌 0

• Achieves state-of-the-art success rates on physically valid pose prediction
• Works across ligand flexibility levels and diverse, unseen protein targets
(4/n)
26.06.2025 11:11 — 👍 1 🔁 0 💬 1 📌 0

PocketVina offers a robust alternative:
• Identifies multiple pocket centers using P2Rank
• Performs GPU-accelerated docking with QuickVina 2-GPU 2.1
• Completes docking + binding affinity prediction in under 1.5 seconds, with no model training
(3/n)
26.06.2025 11:11 — 👍 0 🔁 0 💬 1 📌 0

...physically realistic ligand poses — and are not always as efficient or accurate as often claimed. (2/n)
26.06.2025 11:11 — 👍 1 🔁 0 💬 1 📌 0
I'm excited to share our new preprint: PocketVina — a fast, scalable, and accurate multi-pocket molecular docking method.
Docking remains essential in early-stage drug discovery, but recent deep learning–based approaches still face limitations in generating...
Thread - (1/n)
26.06.2025 11:11 — 👍 4 🔁 4 💬 1 📌 1
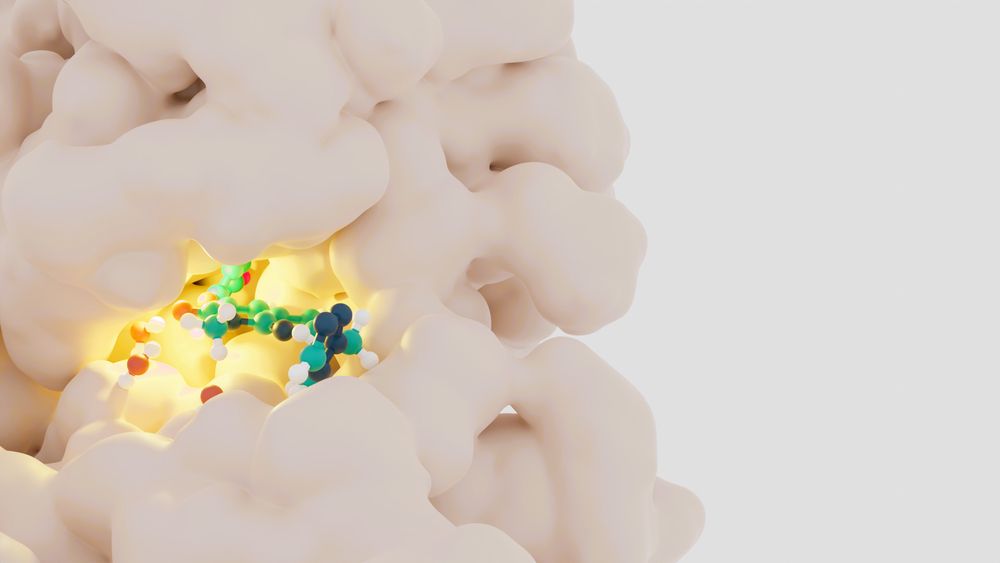
Artistic rendering of a biochemical model: a small molecule ligand, shown as a ball-and-stick model colored by element, is bound in a pocket in a protein surface, shown as a space filling model colored off-white.
We're changing the field of #compchem by creating free and open-source software for performing alchemical free energy calculations. Our flagship protocol calculates relative binding free energies of protein-ligand systems. Try it out in your browser: colab.research.google.com/github/OpenF...
04.02.2025 14:46 — 👍 30 🔁 14 💬 0 📌 0
I remember when I first started learning ML—Andrew Ng offered a Coursera course that uses Octave and covers neural networks for image classification with MNIST. You might find it helpful! :)
03.02.2025 14:26 — 👍 1 🔁 0 💬 1 📌 0

Join us to connect with the vibrant #singlecell community.
📢Register for the #ISCO'25 Conference "Innovations in #SingleCell #OMICS" in Berlin!
🗓️ 12-13 May 2025
🎤 Fantastic Keynote and Invited Speakers
🫵🏿 Many slots for talks: submit your abstract
🔗http://isco-conference.eu
Please spread the word!
31.01.2025 10:29 — 👍 14 🔁 9 💬 1 📌 1
🎓 previously @borklab.bsky.social @EMBL.org & @saezlab.bsky.social @uniHeidelberg.bsky.social
📍Heidelberg
🦠Human microbiome | Host-microbiome interactions | Multiomics integration
PhD candidate at saezlab.bsky.social in computational biomedicine 🔬🧬
Researcher at Heidelberg University @saezlab.bsky.social | #ML and AI in #single-cell and #spatial #omics
Group leader
@sangerinstitute.bsky.social Neural diversity, spatial transcriptomics, glia, GBM
🧬 Structural Bioinformatics | 💊 AI/ML for Drug Discovery | Geometric DL
🔬 @iocbprague.bsky.social, prev. PhD @cusbg.bsky.social @mff.unikarlova.cuni.cz
Computer Science PhD Student at @humboldtuni.bsky.social and @mdc-berlin.bsky.social | Data Science | Machine learning | AI | Bioinformatics | Genomics | Single-Cell Biology
Berlin Institute for Medical Systems Biology
of the Max Delbrück Center @mdc-berlin.bsky.social
www.mdc-berlin.de/en/bimsb
MD-PhD student, computational biology / single-cell omics
Currently at
@cantinilab.bsky.social - Institut Pasteur & @saezlab.bsky.social - Heidelberg Uni
Tools: https://www.github.com/r-trimbour
Publications: https://tinyurl.com/TrimbourRemi
Cofounded and lead PyTorch at Meta. Also dabble in robotics at NYU.
AI is delicious when it is accessible and open-source.
http://soumith.ch
Scientist, Assistant Professor at MIT biology, #FirstGen
Science is the best use of ML
A Nature Research journal on AI, robotics and machine learning
@natureportfolio.bsky.social
nature.com/natmachintell
A @natureportfolio.nature.com journal on mathematical models and computational methods/tools that help advance science in multiple disciplines. https://www.nature.com/natcomputsci
Professor a NYU; Chief AI Scientist at Meta.
Researcher in AI, Machine Learning, Robotics, etc.
ACM Turing Award Laureate.
http://yann.lecun.com
Achira | http://achira.ai
Research laboratory | http://choderalab.org
Antiviral drug discovery for pandemics | http://asapdiscovery.org
OpenADMET | http://openadmet.org
Employer-mandated disclaimer: http://choderalab.org/disclaimer
Pronouns: he/him










