
🧠 The Lipid #Brain Atlas is out now! If you think #lipids are boring and membranes are all the same, prepare to be surprised. Led by @lucafusarbassini.bsky.social with Giovanni D'Angelo's lab, we mapped membrane lipids in the mouse brain at high resolution.
www.biorxiv.org/cgi/content/...
16.10.2025 06:23 — 👍 275 🔁 107 💬 7 📌 11

We are excited to share GPN-Star, a cost-effective, biologically grounded genomic language modeling framework that achieves state-of-the-art performance across a wide range of variant effect prediction tasks relevant to human genetics.
www.biorxiv.org/content/10.1...
(1/n)
22.09.2025 05:29 — 👍 174 🔁 90 💬 4 📌 5
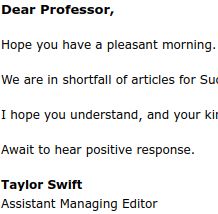
I did not know Taylor Swift was moonlighting in soliciting contributions for fake journals!
16.09.2025 19:29 — 👍 9 🔁 1 💬 0 📌 0
Check out my talented colleagues' study, profiling hundreds of CRISPRa-responsive regulatory elements surrounding PHOX2B, a key player in neuroblastoma, using a targeted scRNA-seq screen in a neuroblastoma cell line.
13.09.2025 16:21 — 👍 1 🔁 0 💬 0 📌 0

Meeting agenda
Sep 10, 2025
Attendees:
Links:
Agenda (feel free to add your items):
• Blog almost ready for R blogger linkage thanks to @Izabela Mamede, @Mengyuan Shen and @Maria Doyle
• New posts from many including
@Juan Henao and myself
• Ideas for other posts?
• There is tidybulk v2 ready to be submitted. Some feedback would be nice there.
• Stefano's new speedy code in tidySE
• https://github.com/tidyomics/
genomics-todos/issues/19#is suecomment-3239791713
• https://github.com/tidyomics/t
idySummarizedExperiment/i ssues/106
• Report back from tidyomics workshop at useR! (Justin and Mike)
• Other projects in the works?
• Ideas for engaging new users?
New developers?
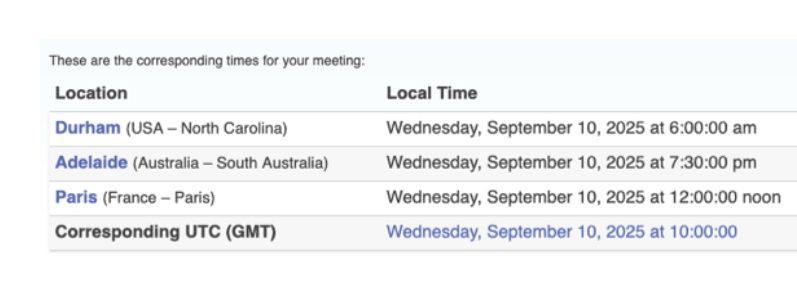
These are the corresponding times for your meeting:
Location
Local Time
Durham (USA - North Carolina)
Wednesday, September 10, 2025 at 6:00:00 am
Adelaide (Australia - South Australia)
Wednesday, September 10, 2025 at 7:30:00 pm
Paris (France - Paris) |
Wednesday, September 10, 2025 at 12:00:00 noon
Corresponding UTC (GMT)
Wednesday, September 10, 2025 at 10:00:00
Our first Fall #tidyomics meeting will be this Wed 10 September, early in US / noon in Europe / late in Australia. Feel free to join if you're interested in what we are doing to make omics data more amenable to tidy data analysis.
Organized with Stefano @stemang.bsky.social
08.09.2025 19:45 — 👍 14 🔁 4 💬 1 📌 1

L’effet Matilda n’est pas une fiction.
Il est inscrit dans l’histoire scientifique.
Il a éclipsé des femmes comme Marthe Gautier, née il y a cent ans, pionnière oubliée de la trisomie 21.
➡️ https://l.franceculture.fr/1LI
10.09.2025 04:00 — 👍 496 🔁 303 💬 9 📌 13

The participants of Dagstuhl Seminar 24122 standing on steps outside (from https://www.dagstuhl.de/24122)

Multiple types of embeddings (UMAP, t-SNE, Laplacian Eigenmaps, PHATE, PCA, MDS) of Wikipedia text data labelled by a text summaries generated by an LLM. Methods like UMAP and t-SNE show cluster structure that reflect shared subject matter in text, whiel other methods show more continuous structure.

Multiple embedding methods (PCA, Laplacian Eigenmaps, t-SNE, MDS, PHATE, UMAP) of primate brain organoids at different time periods. Different methods highlight different aspects of development, such as clusters of similar cell types or time courses of cell development.

Multiple embedding methods (PCA, Laplacian Eigenmaps, t-SNE, MDS, PHATE, UMAP) of 1000 Genomes Project genotypes. Different methods reflect different aspects of demographic history of populations.
Last year I met a bunch of great researchers who work with high-dimensional data at a Dagstuhl seminar. This week we put out a preprint about the history and philosophy of low-dimensional embedding methods, their applications, their challenges, and their possible future arxiv.org/abs/2508.15929
27.08.2025 13:25 — 👍 14 🔁 7 💬 1 📌 1

We spent a year writing this review of low-dim embeddings and arguing about things like epistemic roles and best practices :-) 20+ authors are all participants of the Dagstuhl seminar we held last year: www.dagstuhl.de/24122. Led by @alexandr.bsky.social and Cyril de Bodt.
arxiv.org/abs/2508.15929
27.08.2025 15:14 — 👍 27 🔁 9 💬 1 📌 0
We're committed to support as many attendees as possible join us at #scverse2025 - feel free to reach out if you have questions!
25.08.2025 17:17 — 👍 4 🔁 3 💬 0 📌 0
https://authors.elsevier.com/a/1lbX08YyDfuZWX
Antibodies are highly diverse, but most possible sequences are unstable or polyreactive. In this work, just published in Cell Syst., we propose a new source of data for modeling constraints from these properties. Our models show clear improvements in predicting Ab dysfunction. (1/n)
t.co/qCZERPUMPF
15.08.2025 13:17 — 👍 16 🔁 6 💬 1 📌 0
Thanks, @paubadiam.bsky.social! That makes sense. Excited for the results 🔎.
12.08.2025 18:10 — 👍 0 🔁 0 💬 0 📌 0
Very well set up benchmark and informative comparisons! I might have missed it, but did you also compare the performance of the same methods using either truly paired vs synthetically paired multimodal data as input in terms of your performance evaluation metrics, in addition to network consistency?
12.08.2025 15:11 — 👍 0 🔁 0 💬 1 📌 0
By now, I’ve heard from many people who’ve noticed inconsistencies when using silhouette-based metrics for horizontal data integration evaluation. I hope we’ve helped shed light on why these metrics fall short and that our recommendations prove useful to you!
12.08.2025 07:51 — 👍 4 🔁 0 💬 0 📌 0

Excited to share our latest paper @natmethods.nature.com
We present a high-throughput framework to map cellular interactions at ultra-high scale – broadly applicable from whole-organism immune response mapping to personalized therapy response prediction (1/4).
www.nature.com/articles/s41...
07.08.2025 11:24 — 👍 34 🔁 14 💬 3 📌 0
Lucky to have inspiring and supportive mentors by my side! @mikelove.bsky.social
06.08.2025 08:39 — 👍 1 🔁 0 💬 0 📌 0
Evaluating something like batch correction requires looking at the data, and picking metrics that capture what you care about. Great work @prauten.bsky.social and @uweohler.bsky.social
05.08.2025 12:48 — 👍 25 🔁 8 💬 1 📌 0

Shortcomings of silhouette in single-cell integration benchmarking - Nature Biotechnology
Silhouette score is unsuitable as a metric for single-cell data integration.
Shortcomings of silhouette in single-cell integration benchmarking - @uweohler.bsky.social @prauten.bsky.social @mdc-bimsb.bsky.social @mdc-berlin.bsky.social @humboldtuni.bsky.social go.nature.com/4fcQzZr
30.07.2025 15:26 — 👍 32 🔁 13 💬 1 📌 2
Truly grateful for the exceptional opportunity to participate in #LPSHG2025 last week, featuring a stellar ✨ lineup of leading researchers who doubled as tutors, alongside inspiring fellow PhD students. Excited to apply my learnings and see where this collaborative spirit takes genomics next!
01.08.2025 11:49 — 👍 10 🔁 1 💬 0 📌 0
*Easter egg alert* NOT in the published paper. We also benchmarked Evo 2 and while it did better than other gLMs (consistent that scale can improve gLMs), it still falls short of a basic CNN trained using one-hot sequences and far short of supervised SOTA
16.07.2025 12:16 — 👍 26 🔁 5 💬 1 📌 0
The deadline for the VIB.AI group leader positions is approaching - send in your CV and short research plan before 14th June to start your BioML research lab in Leuven or Ghent
04.06.2025 07:16 — 👍 10 🔁 10 💬 0 📌 0
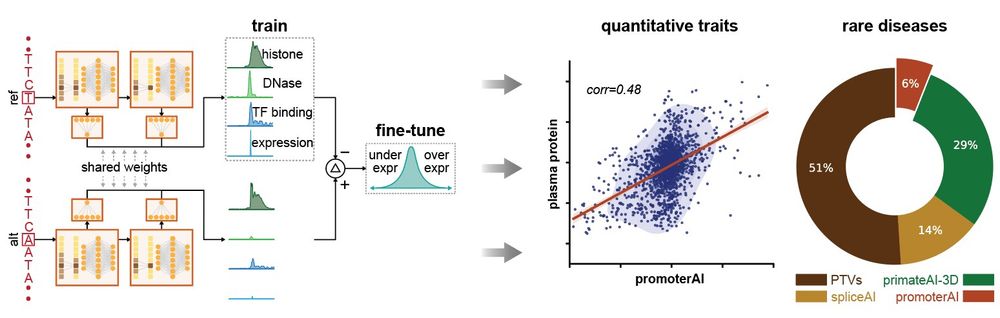
Excited to share my first contribution here at Illumina! We developed PromoterAI, a deep neural network that accurately identifies non-coding promoter variants that disrupt gene expression.🧵 (1/)
29.05.2025 23:57 — 👍 60 🔁 21 💬 1 📌 1


We finally concluded the meeting. Thanks to all attendees for their scientific contributions and for traveling (near or far) to the meeting! Thanks to the local organizers for the infrastructure and catering, and thanks to the co-organizers @yaronorenstein.bsky.social @camillemrcht.bsky.social!
25.04.2025 08:18 — 👍 26 🔁 11 💬 1 📌 0

When investors learn that the trait for green eyes is also ~20 SNPs
14.04.2025 00:45 — 👍 23 🔁 4 💬 0 📌 0
1. LLM-generated code tries to run code from online software packages. Which is normal but
2. The packages don’t exist. Which would normally cause an error but
3. Nefarious people have made malware under the package names that LLMs make up most often. So
4. Now the LLM code points to malware.
12.04.2025 23:43 — 👍 7433 🔁 3381 💬 116 📌 425

40 days until #RECOMB2025 in Seoul! 523 attendees confirmed—thank you! 🥳
For those who haven't registered yet, we have great news!
📢 Early bird deadline is extended to Friday, March 21st 📢
Register now at recomb2025.com 🎟️
15.03.2025 14:08 — 👍 5 🔁 2 💬 0 📌 0
Last day to apply ⏰😱!
If you haven’t already done so, now is the time! Apply to be one of the speakers at the #SoapboxScience summer event and present your research in a fun and relaxed atmosphere ✨
#WomenInSTEM #WomenInScience #Scicomm
21.02.2025 10:25 — 👍 5 🔁 4 💬 0 📌 0

Excerpt from the scikit-learn docs on Manifold learning quoting Baloo's song from the Jungle Book.
Look for the bare necessities with manifold learning... 🐻🎶
Data science meets Disney in the most unexpected places!
Credits to the scikit-learn docs: scikit-learn.org/stable/modul... #DataScience #ManifoldLearning #TechHumor
04.02.2025 13:43 — 👍 2 🔁 0 💬 0 📌 0
Explainable AI research from the machine learning group of Prof. Klaus-Robert Müller at @tuberlin.bsky.social & @bifold.berlin
Official Bluesky account for Berlin Postdoc Days 2025!
https://www.mdc-berlin.de/postdoc-day
Stay tuned for updates for this event! A chance to network with other postdocs working in science in Berlin, career opportunities and a social evening!
Advancing prevention, diagnosis and treatment research for the 30 million people living with a rare disease in Europe.
🔗 erdera.org
Co-funded by European Union's #HorizonEU Research & Innovation programme. Views expressed are of authors only.
Assoc Prof at Karolinska Institutet. Dev bio enthusiast, from liver to brain, and Notch to miRs, cell fates and lineages..
www.anderssonlab.com
We develop machine learning frameworks to decode the geometry and topology of tissue development from spatial omics data | Junior Professor INSERM & Group Leader Turing Center for Living Systems | https://bioml.lis-lab.fr
Statistician and mathematician. Postdoc working on population genetics at the Data Science Institute at Brown University. UWaterloo, Carleton, and McGill alum. Skeets are my own. He/him.
https://github.com/diazale
Translational Immunology Group at Charité - Universitätsmedizin Berlin and Berlin Institute of Health | https://sawitzki-lab.com/
Professor of EECS and Statistics at UC Berkeley. Mathematical and computational biologist.
News from the Genomics of Gene Expression Lab at
@i2sysbio.bsky.social @elcsic.bsky.social. Led by @anaconesa.bsky.social
Computer scientist at I2SysBio-CSIC. Transcriptomics, long-reads, multiomics, tool developer. Promoter of Green Science. Finally happy!
Account of the Saez-Rodriguez lab at EMBL-EBI and Heidelberg University. We integrate #omics data with mechanistic molecular knowledge into #opensource #ML methods
Website: https://saezlab.org/
GitHub: https://github.com/saezlab/
European 🇪🇺 | #Berlin 🏠 | PhD student in medical systems biology 🧬 with @leifludwig.bsky.social | Cooking & mountains
Postdoctoral researcher at @anshulkundaje.bsky.social Machine learning #ML, gene regulatory networks #GRN, single cell and spatial #omics.
Previously at @saezlab.bsky.social
Core developer of https://decoupler.readthedocs.io/ at @scverse.bsky.social
Postdoc in the Transcriptomics and Functional Genomics Lab at the Barcelona Supercomputing Center studying the impact of human variation on the transcriptome. California -> Catalunya
Computational Pathology Biomarker Lead at AstraZeneca | PhD
Likes immune cells, single-cell technologies, and home made pizza
Postdoc // gene regulation, genomics, synbio, stem cells, c. elegans // with @eddaschulz @molgen.mpg.de previously @n_rajewsky @mdc-bimsb //
Cells talk; I build tools to listen to them
Computational biologist • Occasional composer and pianist • he/him
Opinions are my own.
Assistant Professor 👩🏼💻 at Erasmus Medical Center Rotterdam researching genetics & gene regulation 🧬🖥 in the context of disease 🥼💉📊






















