📢 @bayraktarlab.bsky.social & colabs at @sangerinstitute.bsky.social are assembling the @opentargets.org funded Neuroimmune Team 🧠🧬-Leverage AI to uncover neurodegeneration
Tech Spec: sanger.wd103.myworkdayjobs.com/en-US/Wellco...
Postdoc AI: sanger.wd103.myworkdayjobs.com/en-US/Wellco...
30.06.2025 09:07 — 👍 1 🔁 2 💬 0 📌 0
Very excited about our new chapter with @mhaniffa.bsky.social !
24.05.2025 07:14 — 👍 6 🔁 0 💬 0 📌 0
Thanks Muzz!
18.05.2025 10:23 — 👍 0 🔁 0 💬 0 📌 0
Thanks Adrien!
18.05.2025 10:23 — 👍 1 🔁 0 💬 0 📌 0
Thanks Chris!
17.05.2025 08:08 — 👍 0 🔁 0 💬 0 📌 0
Grateful to >60 authors for their amazing work + @wellcomeleap.bsky.social (special thx to Jason Swedlow for leading DeltaT!) & @cziscience.bsky.social for their support. @ebi.embl.org @crick.ac.uk @cambridgeuni.bsky.social @dkfz.bsky.social @sangerinstitute.bsky.social
16.05.2025 11:42 — 👍 1 🔁 0 💬 0 📌 0
There are many implications & LOTS of detail in the preprints (e.g. mesenchymal cancer states is really a gliosis/hypoxia response & GB tumour subtypes are likely confounded) bit.ly/4mkrWgs bit.ly/3FbI6Ic
16.05.2025 11:42 — 👍 1 🔁 0 💬 1 📌 0
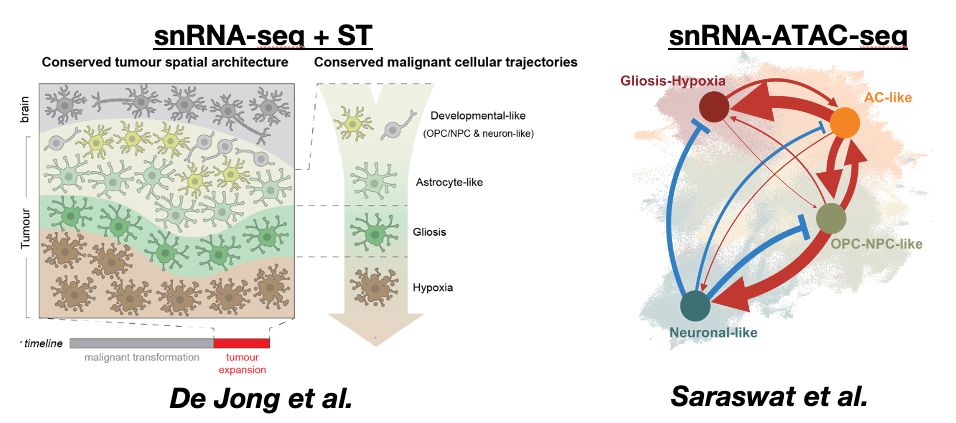
Taken together, these 2 studies start to unveil what i call the “rules” of glioblastoma. Despite their incredible complexity, these tumours are governed by specific cellular & tissue mechanisms. We believe/hope this understanding will be a foundation for new therapies
16.05.2025 11:42 — 👍 2 🔁 0 💬 1 📌 0
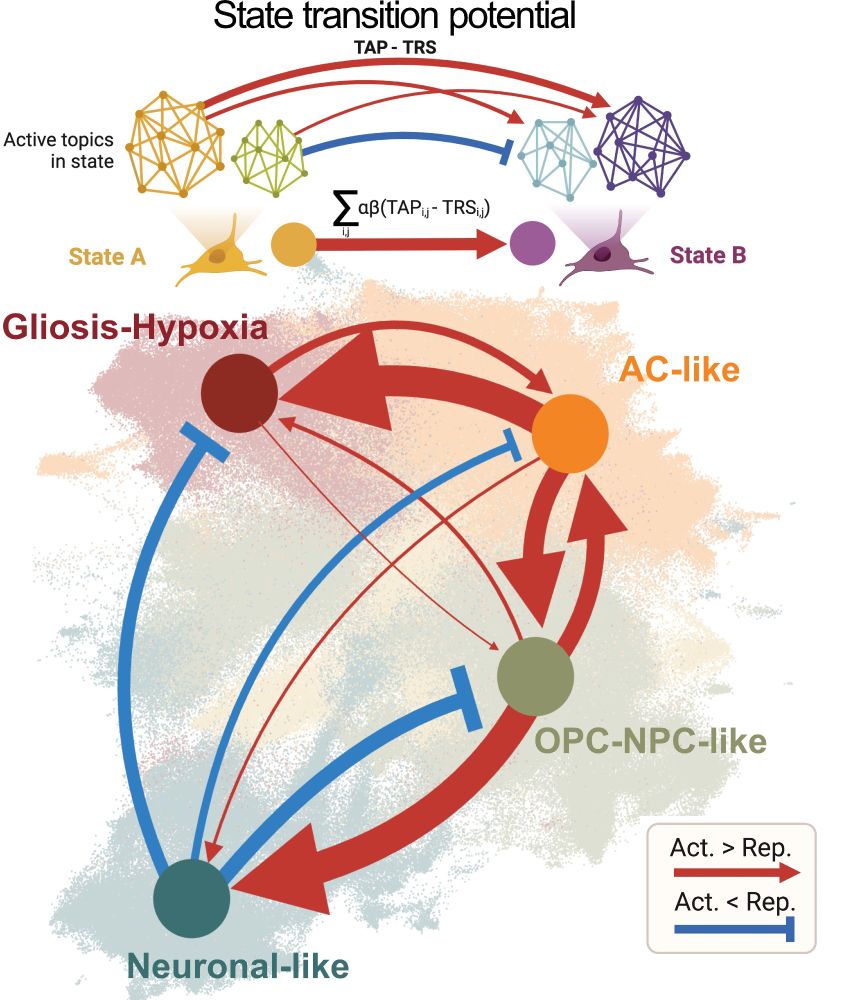
In GB, as opposed to unrestrained plasticity of cancer cells, we predict transition highways and barriers aligned with paper 1. And we mechanistically validate a transcriptional repressor safeguarding neuronal like cancer states!
16.05.2025 11:42 — 👍 1 🔁 0 💬 1 📌 0
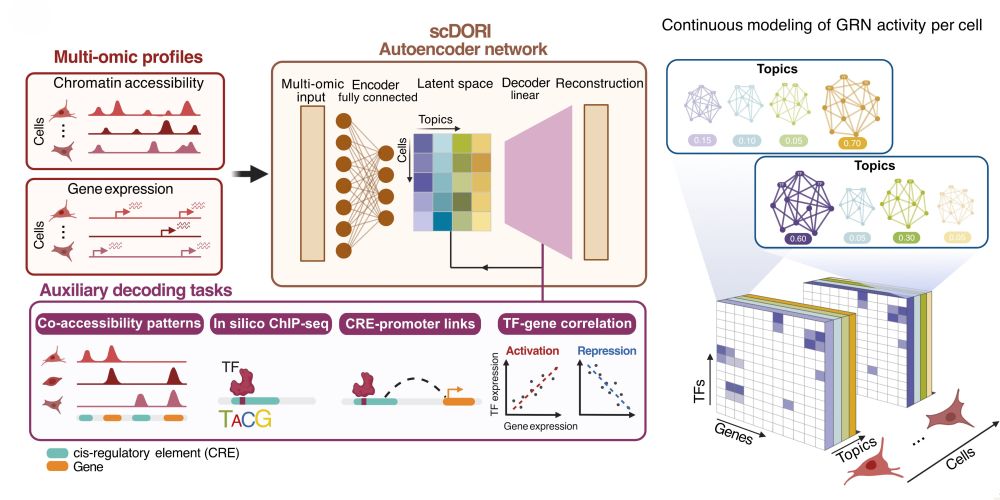
scDORI enables us to examine GRNs in a continuous manner across cellular trajectories. Hence, you don’t just map GRNs / TF regulators, you can actually infer cell plasticity and predict the future state of a given cell
16.05.2025 11:42 — 👍 1 🔁 0 💬 1 📌 0
Here we developed a new deep learning based GRN inference framework: scDORI. See the thread from Manu for more details but few highlights here bsky.app/profile/manu...
16.05.2025 11:42 — 👍 3 🔁 0 💬 1 📌 0
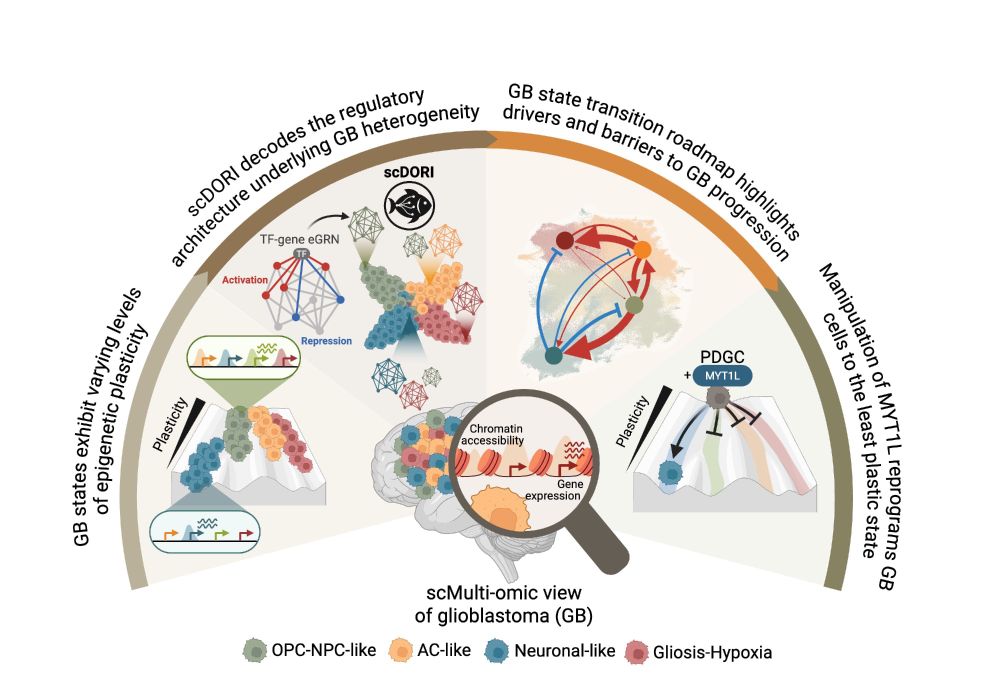
In paper 2, we leverage our massive snRNA-ATACseq data to predict cancer cell state transitions & trajectories from gene regulatory networks. Led in @oliverstegle.bsky.social and Moritz Mall's labs by @manusaraswat.bsky.social @lauraruedag.bsky.social Elisa + Tannia & Fani
16.05.2025 11:42 — 👍 1 🔁 0 💬 1 📌 0
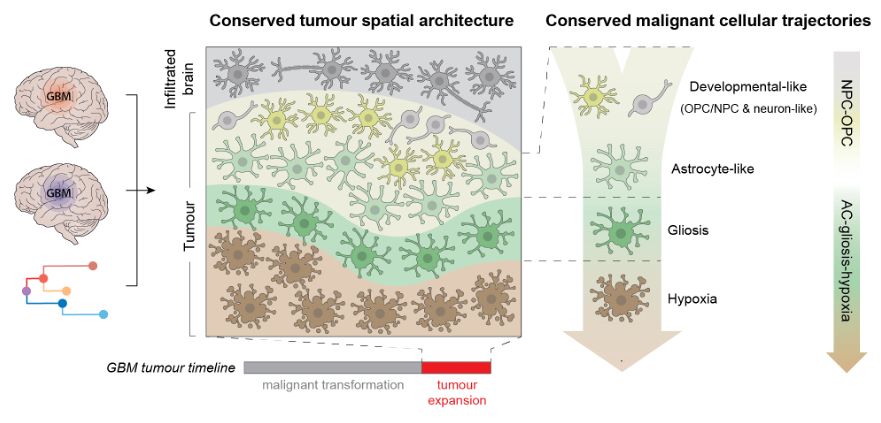
Hence, we define a stereotyped trajectory of cancer cells at the heart of GB heterogeneity - unifying previously defined cell states & tumour subtypes
16.05.2025 11:42 — 👍 1 🔁 0 💬 1 📌 0
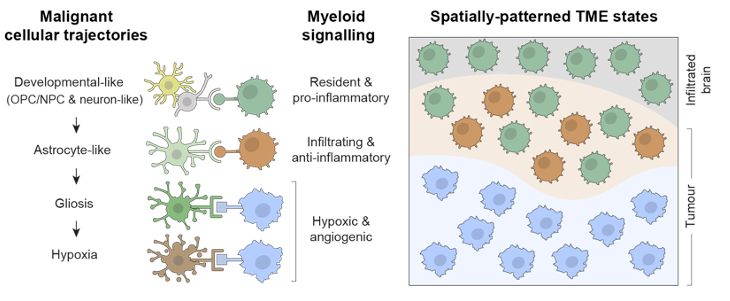
Finally, cancer cells are not riding alone. We found that this cancer cell trajectory is intimately linked to myeloid heterogeneity and unfolds across regionalised myeloid signalling environments.
16.05.2025 11:42 — 👍 1 🔁 0 💬 1 📌 0
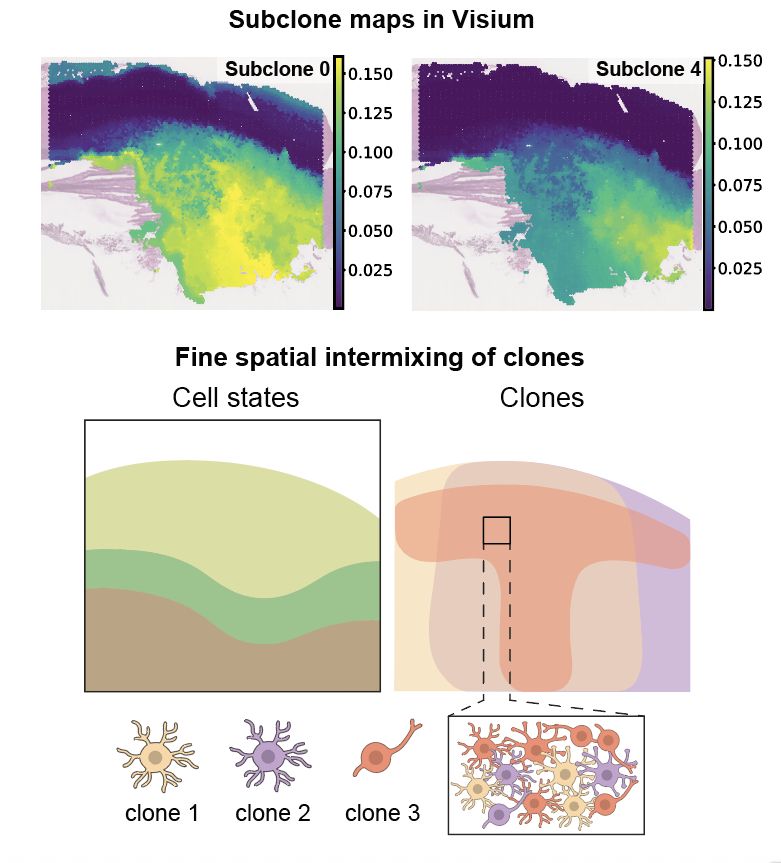
This led to a wonderful side quest: we mapped tumour subclones in space! We developed SpaceTree to deconvolve tumour subclones in Visium data and found that they are finely spatially intermixed (within Visium spots) in the tissue! github.com/PMBio/spaceT...
16.05.2025 11:42 — 👍 3 🔁 0 💬 1 📌 0
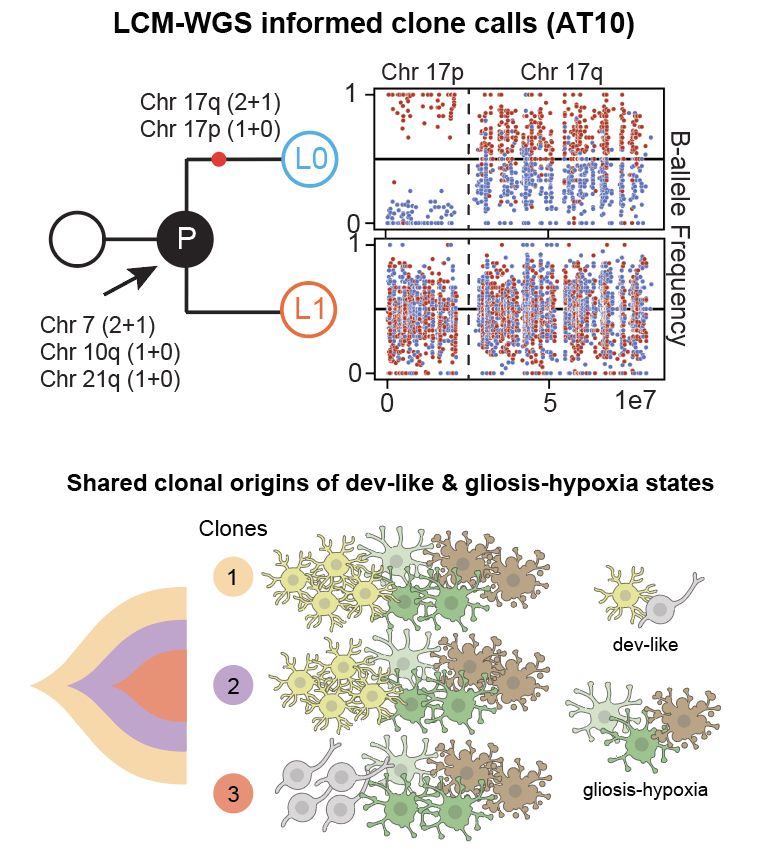
These results suggest that this trajectory cuts across the genetic hierarchy of GB i.e. shared across tumours/subclones. We used spatial DNA-seq (LCM-WGS) to validate this +found the trajectory is shared, yet genetically malleable.
16.05.2025 11:42 — 👍 2 🔁 0 💬 1 📌 0
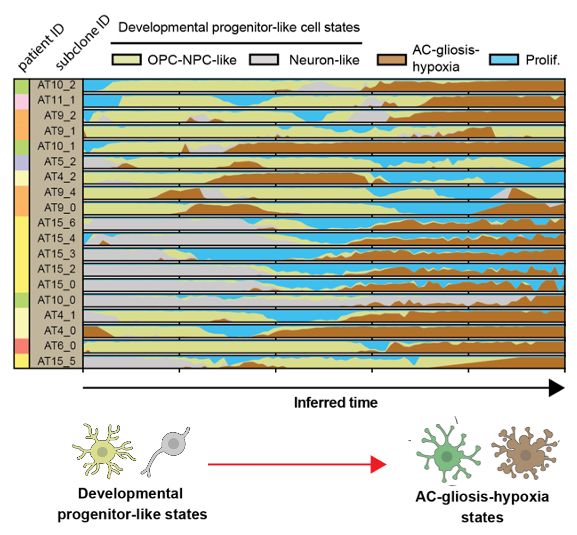
Unbiased transcriptomic trajectory analysis (i.e. our cell2fate RNA velocity go.nature.com/4km62aI) finds that this is trajectory shared across tumours + genetic subclones. And we define a molecular progression conserved across tumours/clones
16.05.2025 11:42 — 👍 3 🔁 0 💬 1 📌 0
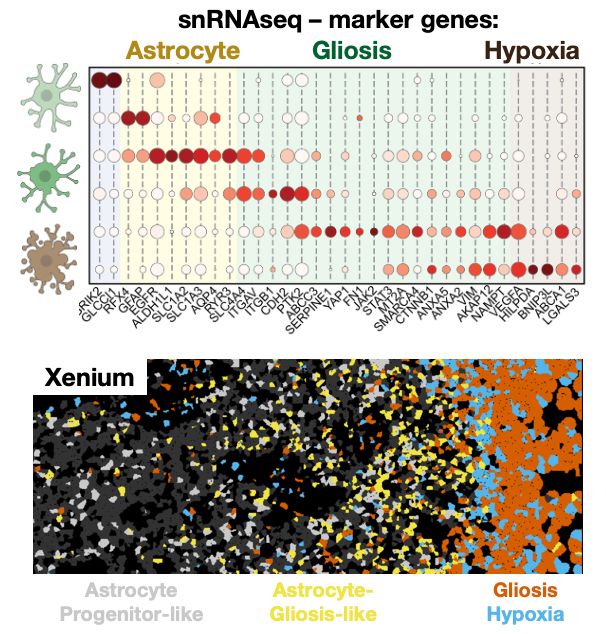
When you look into the tumour areas/states dominated by gliosis/hypoxia, these transitions become clear. You can see cancer cells travelling from developmental astrocyte like states gradually into gliosis + hypoxia response (more on this below)
16.05.2025 11:42 — 👍 5 🔁 0 💬 1 📌 0
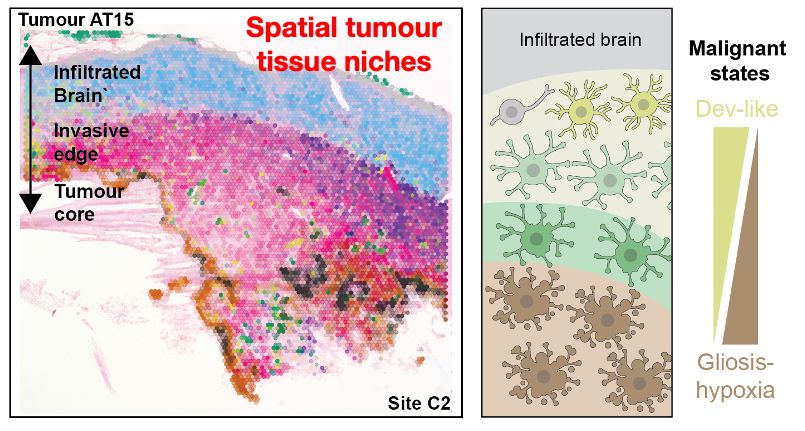
..to resolving this trajectory. Integrating single nuclei + spatial data revealed stepwise spatial transitions of cancer cell states from the tumour invasive edge towards the necrotic core. This looked like spatially organised cancer cell transitions as GB tumours rapidly expand.
16.05.2025 11:42 — 👍 3 🔁 0 💬 1 📌 0
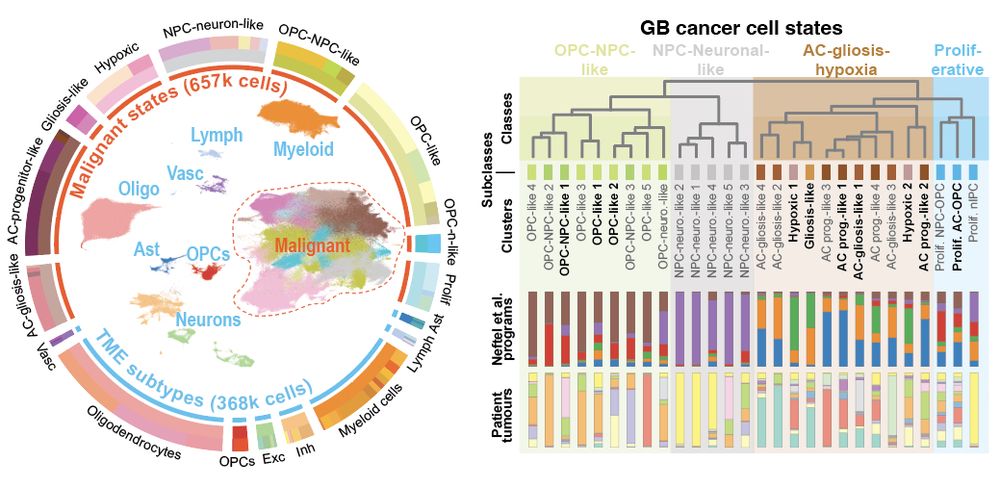
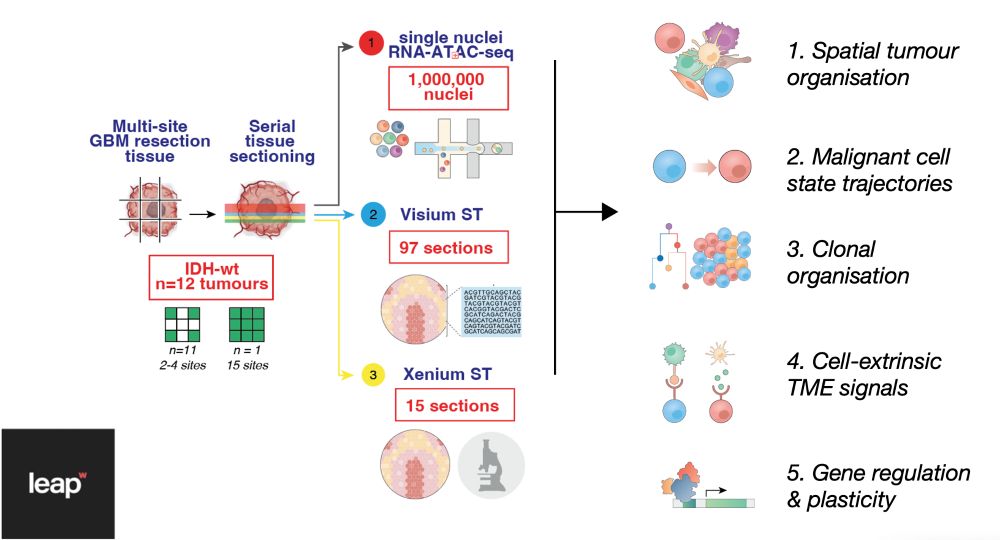
It started with single nuclei RNA-seq. Profiling >1 million cells, we saw within each tumour that cancer cells vary from developmental progenitor-like states (e.g.NPC/OPC) to those marked by gliosis (i.e. glial injury response) & hypoxia Yet, the spatial/tissue context was the key…
16.05.2025 11:42 — 👍 2 🔁 0 💬 1 📌 0
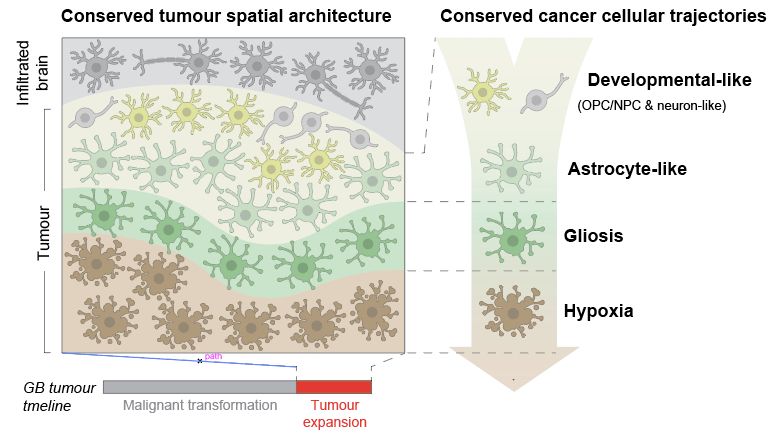
Integrating these modalities, we interpreted human GB biology from scratch. We mapped GB heterogeneity to a spatially-patterned trajectory of cancer cells in tissue, where they transition from developmental-like towards glial injury response & hypoxia states.
16.05.2025 11:42 — 👍 2 🔁 0 💬 1 📌 0

In paper 1, we tackled this big question. We created the GBM-space, the most comprehensive atlas of human GB. It uniquely pairs single cell + spatial modalities to bridge the cell, tissue and genetic basis of GB. Led by the amazing team of Grant, Tannia, Fani & Olga
16.05.2025 11:42 — 👍 2 🔁 0 💬 1 📌 0
BUT despite elegant work, we still don’t know how/why cancer cells diversify & transition across states. Do cancer cells follow a defined trajectory across tumours? Or is it a chaos of unrestrained plasticity? Treatments will continue to fail if we lack this basic understanding
16.05.2025 11:42 — 👍 2 🔁 0 💬 1 📌 0
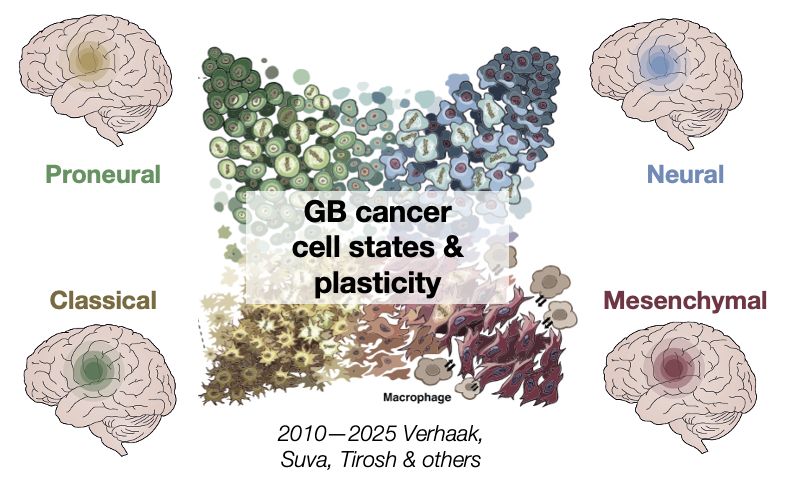
Glioblastoma (GB) is a rapidly fatal brain tumour. Within each GB tumour, cancer cells exist across diverse cellular states + can switch their states (i.e. plasticity) + are thought to make tumour subtypes….
16.05.2025 11:42 — 👍 2 🔁 0 💬 1 📌 0

Over the past 3 years, we led a fantastic @wellcomeleap.bsky.social collaboration w. @oliverstegle.bsky.social, Behjati, Mall, Mair, @jamesbriscoe.bsky.social & @juliosaezrod.bsky.social labs. We created a massive multiomic atlas & new comp models to discover human GB biology www.gbmspace.org
16.05.2025 11:42 — 👍 16 🔁 6 💬 1 📌 1
Scientist developing AI for oncology. Division head at the German Cancer Research Centre DKFZ. Prof at the University of Heidelberg, Germany. Previously at EMBL-EBI and Wellcome Sanger Institute. Alumnus of ETH Zurich.
Group Leader @umeauniversitet.bsky.social
Wallenberg Fellow in Molecular Medicine at WCMM Umeå
Postdoc @embl.org
PhD @cniostopcancer.bsky.social
3D genomics , epigenomics, gene regulation, enhancer, cancer, glioblastoma, cancer neuroscience
Assistant Professor @YaleMed | tissue biology, computation, systems biology |
www.hattiechunglab.bio
Group leader @schapirolab.bsky.social at the Heidelberg University Hospital focusing on #HighlyMultiplexedImaging and #spatialomics #spatialbiology technologies and analysis. President of the European Society for Spatial Biology e.V. (ESSB)
Senior Editor @Nature for cancer and cell cycle. Views my own
Cancer Researcher at Princess Margaret Cancer Centre, Assistant Professor at U of T
Stem Cell and Cancer Biologist | Lecturer | PI | @ucl.ac.uk | @ucllifesciences.bsky.social | School of Pharmacy
Junior research group leader and clinician scientist @Uniklinik Heidelberg&DKFZ. Epigenetics in brain tumors.
We study mechanisms of cancer progression and metastasis - focus on colorectal cancer
Group Leader and Wellcome Trust Career Development Fellow at the UK Dementia Research Institute at Edinburgh University. Interested in astrocytes at brain borders.
Chileno 🇨🇱 - Postdoc at the Wellcome Sanger Institute. PhD in Bioinformatics & Systems Biology from UC San Diego.
https://earmingol.github.io
Studying the spatial organization of brain tumors @ the Tirosh Lab @ the Weizmann Institute. Mother of 4. Lover of candy, art, & books.
PhD student in the Saez-Rodriguez group (@saezlab.bsky.social) @ebi.embl.org. Member of Gonville & Caius College (@caiuscollege.bsky.social).
Computational biology | Omics | Microbiome | IBD & CRC | Open science
PhD candidate at German Cancer Research Center (DKFZ)
Researcher at Heidelberg University @saezlab.bsky.social | #ML and AI in #single-cell and #spatial #omics
Research group leader @German Cancer Research Center, reconstructing human development and disease using cell fate engineering
Statistician and computational biologist; uw alum; jhu student. He/him. http://ekernf01.github.io
Assistant Professor of Pathology @yalecancer.bsky.social • @cancerresearchuk.org Clinician Scientist & Pathologist @cam.ac.uk • www.aitkenlab.org • #GenomicPathology • posts≠employer •
🚲 https://www.rideclosertofree.org/participants/SarahAitken
Wellcome Trust Clinical PhD Fellow in the Garnett Lab @sangerinstitute.bsky.social. Interested in cancer plasticity, multiplexed assays, ML, & comp bio.
https://ttszen.com/