Now published: our work using phylodynamics from surveillance data to quantify and experimentally validate the fitness impact of antibiotic resistance determinants & how this changes with patterns of antibiotic use: www.nature.com/articles/s41...
30.01.2026 12:34 — 👍 38 🔁 17 💬 1 📌 0
New paper out in @pnas.org, and it made the cover! 👁️
We represent plasmids as circles and mutations as dots, resembling an eye, because in this paper we literally 𝑤𝑎𝑡𝑐ℎ plasmids evolve.
‼️Check Paula’s 🧵 and the paper👇
𝗣𝗹𝗮𝘀𝗺𝗶𝗱 𝗺𝘂𝘁𝗮𝘁𝗶𝗼𝗻 𝗿𝗮𝘁𝗲𝘀 𝘀𝗰𝗮𝗹𝗲 𝘄𝗶𝘁𝗵 𝗰𝗼𝗽𝘆 𝗻𝘂𝗺𝗯𝗲𝗿
www.pnas.org/doi/10.1073/...
27.01.2026 20:23 — 👍 96 🔁 44 💬 3 📌 3
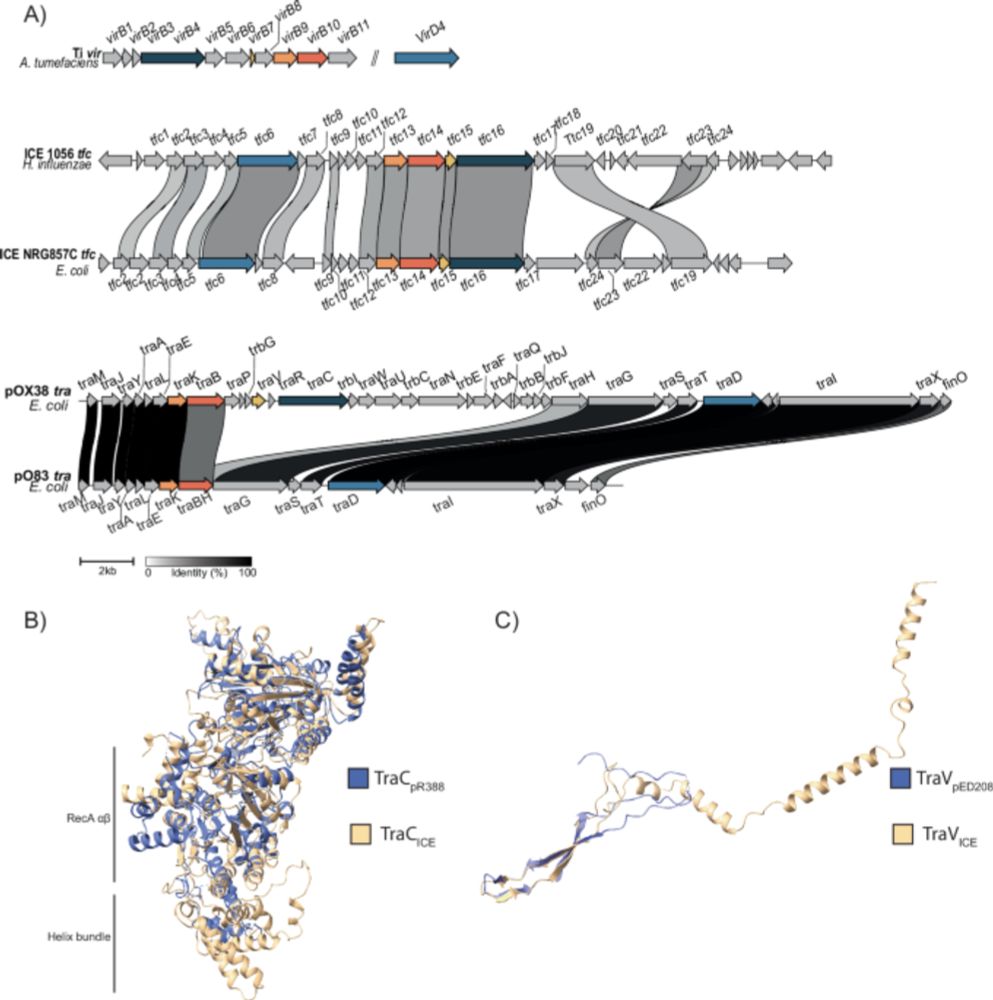
Bacteria chromosomes contain Genomic Islands that provide virulence, antibiotic resistance, MGE-defence,... They transfer between cells, but the mechanism of most remains elusive.
Here we explore the conjugative capacity of these mysterious Genomic Islands.
www.biorxiv.org/content/10.6...
14.01.2026 10:14 — 👍 82 🔁 52 💬 4 📌 2

What drives the K. pneumoniae species complex (KpSC) to thrive from hospitals to soils? In this study, we investigate how pangenome structure and functional diversity shape KpSC adaptability across phylogenetic and ecological contexts. (Thread)
www.biorxiv.org/content/10.6...
16.12.2025 11:59 — 👍 9 🔁 3 💬 1 📌 1
A new tool we’ve developed to identify AMR-associated SNPs in short- and long-read metagenome data….allows a greater understanding of the total resistome of a sample
08.12.2025 10:51 — 👍 9 🔁 3 💬 0 📌 0

🧬 A new “Life Identification Number (LIN)” system aims to modernize bacterial classification with a clearer, more stable hierarchy. A step forward for genomic taxonomy!
📖 shorturl.at/vkQuS
✍️ @sylvainbrisse.bsky.social & coll.
@pasteur.fr @ox.ac.uk @monashuniversity.bsky.social @lshtm.bsky.social
30.11.2025 08:20 — 👍 8 🔁 4 💬 1 📌 0
2026-27 Project (Dyson & Mostowy & Knight) - MRC London Intercollegiate Doctoral Training Partnership Studentships
PHACTS: Unravelling PHAge-baCTeria-host immune dynamicS to inform phage therapy SUPERVISORY TEAM Supervisor Dr Zoe Dyson...
London interdisciplinary #PhD position now open with myself, @sergemostowylab.bsky.social, & @gmknght.bsky.social on #phage -bacteria-host immune dynamics for WHO priority bacterial pathogens #Klebsiella, #Shigella, and #Staph. Combines cellular microbiology, genomics, & mathematical modelling.
13.11.2025 11:11 — 👍 9 🔁 12 💬 0 📌 0

🧬 New #GenEpiBioTrain course!
Bacterial Strain Taxonomy for Genomic Surveillance
📅 22–23.10, 09:00–12:00 (CEST)
Learn how bacterial pathogens are classified & named in genomic surveillance – from #MLST & #cgMLST to SNP-based & k-mer clustering.
Info: bit.ly/48qT5df
#ECDCTraining #IDsky #EpiSky
08.10.2025 14:33 — 👍 8 🔁 7 💬 2 📌 0
Dear community, Bakta needs your help!
To further improve the functional annotation of "hypothetical" CDS, me and @gbouras13.bsky.social, we are looking for the worst Bakta-annotated bacterial genomes ;-)
(1/2)
06.10.2025 07:27 — 👍 11 🔁 18 💬 1 📌 0
Now published, our tool to run (almost) all biological models interactively in your web browser
Paper: academic.oup.com/bioinformati...
Website: biomodels.bacpop.org
Code: github.com/bacpop/SBMLt...
29.09.2025 09:49 — 👍 18 🔁 9 💬 1 📌 0
To summarise our recent pre-print: Autocycler, the automated consensus assembler, when used with Nanopore long-read only Enterobacterales assemblies, produces more complete chromosomes and plasmids, with an accuracy comparable to hybrid assemblies.
29.09.2025 06:57 — 👍 26 🔁 16 💬 1 📌 0

Delighted to see our paper studying the evolution of plasmids over the last 100 years, now out! Years of work by Adrian Cazares, also Nick Thomson @sangerinstitute.bsky.social - this version much improved over the preprint. Final version should be open access, apols.
Thread 1/n
25.09.2025 21:28 — 👍 299 🔁 154 💬 14 📌 8
EMBL International PhD Programme – Unique in the world and waiting for you!
The EMBL PhD programme is open until 13th October (entry ~Sep 2026):
www.embl.org/about/info/e...
We have three positions in microbial genomics at EMBL-EBI, including one in my group. Please do apply, or if you know anyone that would be interested pass on to them
17.09.2025 15:49 — 👍 19 🔁 39 💬 1 📌 3
Thanks for all the trainers and attendees on this course, which was a lot of fun to run, and hopefully filled a gap in genomics/modelling training
And especially co-organisers @leonielorenz.bsky.social @sonjalehtinen.bsky.social Joel Hellewell
22.09.2025 09:43 — 👍 12 🔁 4 💬 1 📌 0
Open PhD position in our lab for starting roughly in September 2026. Cannot recommend this lab enough!!! 🦠🖥️
#MathematicalModelling
#BacterialGenomics
#Bioinformatics
19.09.2025 13:37 — 👍 3 🔁 1 💬 0 📌 0
GitHub - samhorsfield96/ExpEvoAnalyzer: A workflow to analyse experimental evolution data.
A workflow to analyse experimental evolution data. - samhorsfield96/ExpEvoAnalyzer
A little tool I've developed: ExpEvoAnalyzer (github.com/samhorsfield...) - a snakemake pipeline that compares isolate paired-read data from an experimental evolution study to a reference isolate, producing functionally-annotated SNPs in a presence/absence matrix.
11.09.2025 14:16 — 👍 8 🔁 2 💬 1 📌 0
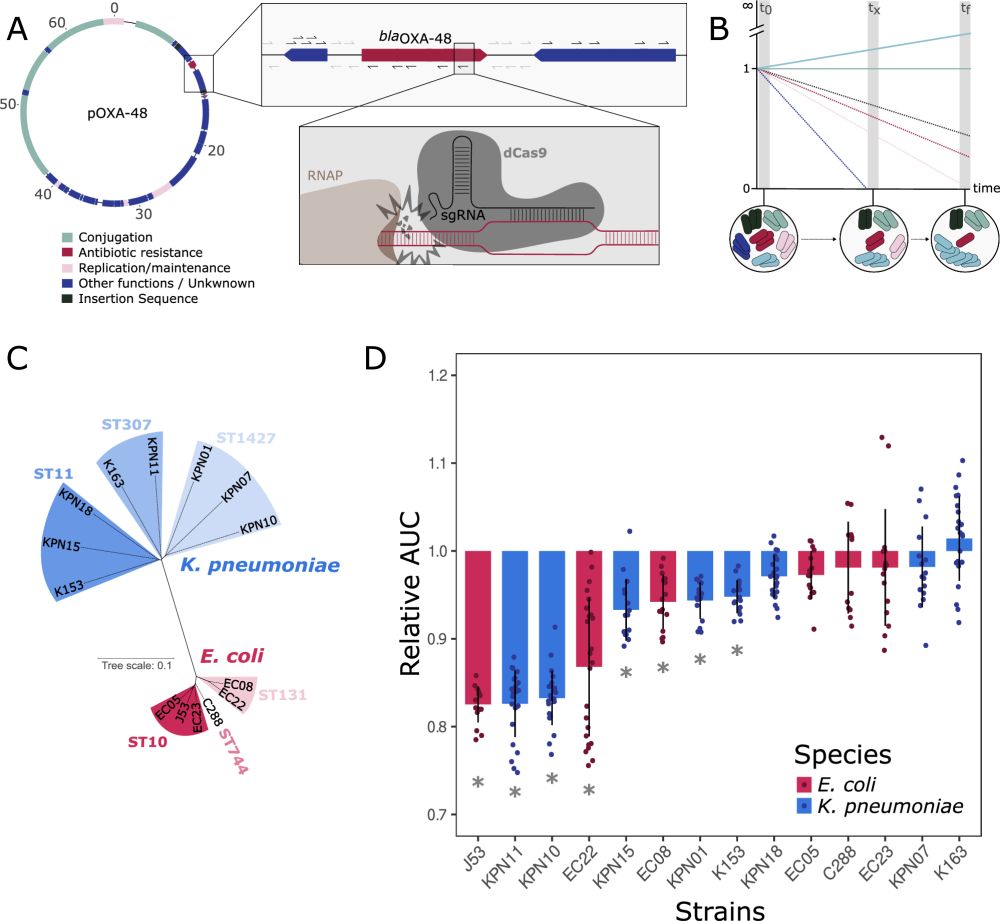
Dissecting pOXA-48 fitness effects in clinical Enterobacterales using plasmid-wide CRISPRi screens
Nature Communications - This study investigates the effects of the carbapenem resistance plasmid pOXA-48 in clinical enterobacteria. Using CRISPRi screens, the authors revealed that the...
This work is finally published! 🥳🧬
Plasmids are associated with very variable fitness costs in their different bacterial hosts. But, what is the contribution of each of the plasmid-genes in these host-specific effects? Study led by
@jorgesastred.bsky.social, @sanmillan.bsky.social and myself! 1/14
20.08.2025 13:25 — 👍 82 🔁 41 💬 6 📌 5
LGBTQ+ genome scientist, microbiologist... give me a cool genome structure & I am happy! Proud member of @langridgelab.bsky.social & creator of #WhatIsAScientist exhibition
University of Aveiro
https://pratas.github.io
Bioinformatics
Microbial ecology and evolution 🧬
Postdoc @pasteur.fr GEM
PhD @inrae-maiage.bsky.social
Executive Publisher at T&F. Publish journals. Walk dogs. ❤️ doughnuts & ice cream. #scicomm #physicalsciences #mathematics #statistics #datascience #history #science #STS
PhD candidate at UW Seattle. I study how interactions between viruses shapes their evolution. Also 📷, 🎹, 📽️, 🐦
MD PhD. Interested in infectious diseases, biofilms, bacterial evolution, adaptation and AMR.
PhD candidate in Medical Microbiology, University of Zurich | AMR evolution in the gut microbiome
NIHR Clinical Lecturer at Imperial College London | Infectious Diseases and Microbiology Doctor | PhD at Wellcome Sanger Institute and University of Cambridge
Research Fellow in van Schaik Lab, University of Birmingham. AMR, functional genomics, evolution, drug discovery
ESCMID Study Group for mobile elements and plasmids
Computational microbiologist. Senior scientist at @cemess.bsky.social, @univie.ac.at.
Microbial ecology, mostly of nitrogen cycle microbes, and data driven physiology.
Maintainer of the GlobDB genome database https://globdb.org
Mom of two ❤️
Early-career researcher in AMR, Mobile Genetic Elements, and One Health & Part-time lecturer.
In love with the intelligence of microbes, their evolutionary designs, and how they outthink every threat.
JUST- Jordan 🇯🇴
assistant professor at ucsf interested in genetics, statistics, etc…
jeffspence.github.io
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, AI/ML in biotech // http://albertvilella.substack.com
Lecturer in Microbiology at the University of Reading. Interested in uncovering how bacteria fight it out and using those strategies against them
Senior Postdoc @enningalab.bsky.social, @pasteur.fr, always working on Salmonella host-pathogen interactions | Bookworm | Enjoyer of many fictional worlds | Mother of Cats | She/her 🩷💜💙
PhD in Biology (Génétique et Génomique)
Evolution of integrons, in the lab of @epcrocha.bsky.social, Microbial evolutionary genomics @pasteur.fr
@EPFL and @Polytechnique alumni
La resistencia a los antimicrobianos representa una amenaza creciente para la sociedad. En AMR Hub CSIC impulsamos soluciones innovadoras para frenarla y garantizar la eficacia de los antibióticos, tanto hoy como en el futuro.
Bacterial Mobile Evolution. Staff Scientist at Sanger Institute. I study how pathogens emerge and evolve by sharing genes. Interested in AMR, Plasmids, Phages, ICEs, and everything that moves within and between genomes.
We are a world leader in genomics research. We apply and explore genomic technologies to advance the understanding of biology and improve health 🧬
https://www.sanger.ac.uk/