
Efficient sequence alignment against millions of prokaryotic genomes with LexicMap - Nature Biotechnology
LexicMap uses a fixed set of probes to efficiently query gene sequences for fast and low-memory alignment.
Sometimes you meet absolutely incredible bioinfo-magicians.
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...
10.09.2025 09:12 — 👍 188 🔁 99 💬 5 📌 4

How do bicycles work?
Conservation of angular momentum plays a major role, but why is angular momentum conserved?
A post about Emmy Noether, one of the most influential mathematical physicists of the 20th century, who rarely gets the recognition she deserves.
blog.genesmindsmachines.com/p/how-do-bic...
16.09.2025 14:03 — 👍 8 🔁 3 💬 0 📌 0
Where are the ubam nowadays? GATK promoted them long time ago
17.09.2025 14:42 — 👍 0 🔁 0 💬 0 📌 0

Over the next few months, we will be hosting a Workflow Demo Series to cover everything you'll need to know as you explore Positron firsthand.
📆 First session: Getting Started with Positron: Quick Tour and Community Q&A on September 24th! Register here - events.zoom.us/ev/Ajss5j9Ve...
16.09.2025 17:47 — 👍 10 🔁 6 💬 0 📌 0

If all the world were a monorepo
The R ecosystem and the case for extreme empathy in software maintenance
Really insightful post from Julie Tibshirani (spotted in LinkedIn, can't find on Bsky) reflecting on #rstats 's unique governance structure and what can be learned for other languages
jtibs.substack.com/p/if-all-the...
14.09.2025 23:29 — 👍 126 🔁 50 💬 7 📌 8
Naughty Grok must be taught not to tell the truth
14.09.2025 21:18 — 👍 4 🔁 3 💬 0 📌 0
YouTube video by Senator Bernie Sanders
In Wake of Charlie Kirk Murder, Sen. Bernie Sanders Addresses Rising Political Violence in America
How to effectively talk about political violence. Bernie Sanders nailed it again
youtu.be/HlIvH6ozvv4?...
13.09.2025 18:06 — 👍 1 🔁 0 💬 0 📌 0
Snakemake
#Snakemake 9.10 has been released. The major change is that it now supports scheduling plugins. By that your scheduling algorithm research becomes immediately usable with thousands of workflows by thousands of users. snakemake.github.io
29.08.2025 12:54 — 👍 7 🔁 5 💬 0 📌 0
Hey friends! Are you interested in the science of learning? Then check out all the cool SOL researchers in this starter pack! go.bsky.app/CBPm1dV
#academicsky #edusky #psychsky
23.08.2025 13:24 — 👍 35 🔁 15 💬 2 📌 1
Google
Sign in
is dunkin cereal milk gluten fr
Al Overview
+1
No, Dunkin' Cereal Milk is not gluten-free because it contains milk, which is a common allergen, and as a result, it is not suitable for those with celiac disease or gluten intolerance.
However, coffee drinks, dairy milk, and non-dairy milks like oat, almond, and coconut milk are naturally gluten-free at Dunkin'.
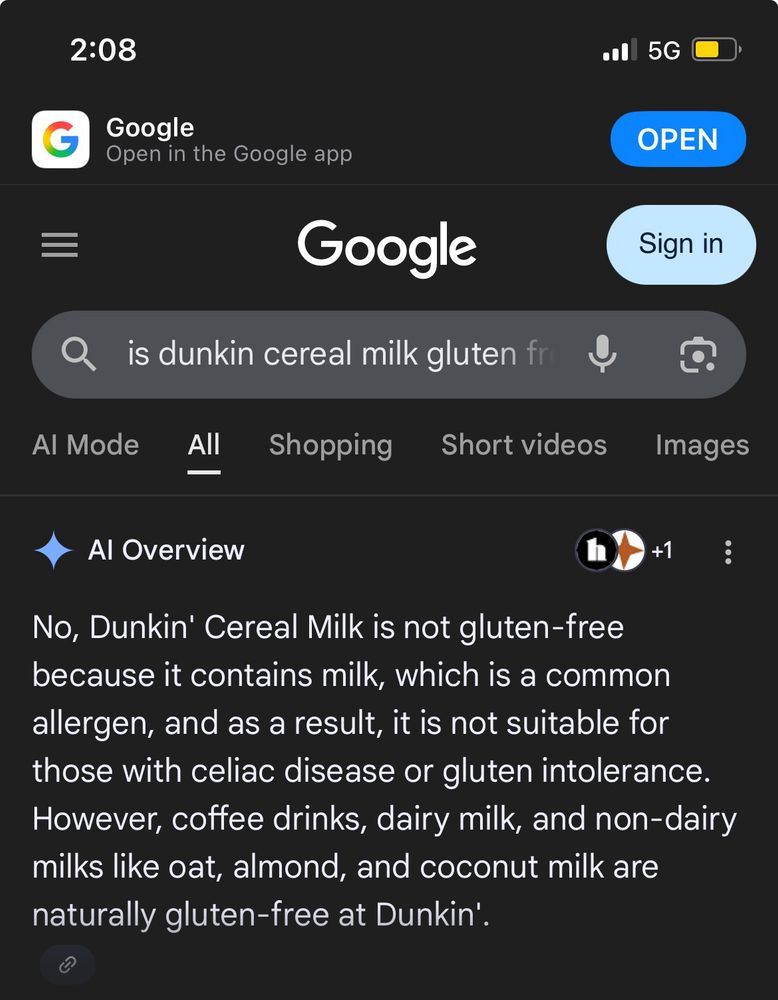
In 25 years, every business school in the country will be doing case studies about how a long defunct company known as “Google” once had an unbeatable lock on online information retrieval and then started doing shit like this.
23.08.2025 18:16 — 👍 8426 🔁 2001 💬 237 📌 134
🚨 Imagine a bacterium that refuses to follow the textbook:
It grows as tangled filaments, divides unevenly, reshapes its own membranes… and even builds grappling hooks.
Meet Litorilinea aerophila — and here’s why it blew our minds.
20.08.2025 10:34 — 👍 166 🔁 70 💬 5 📌 5
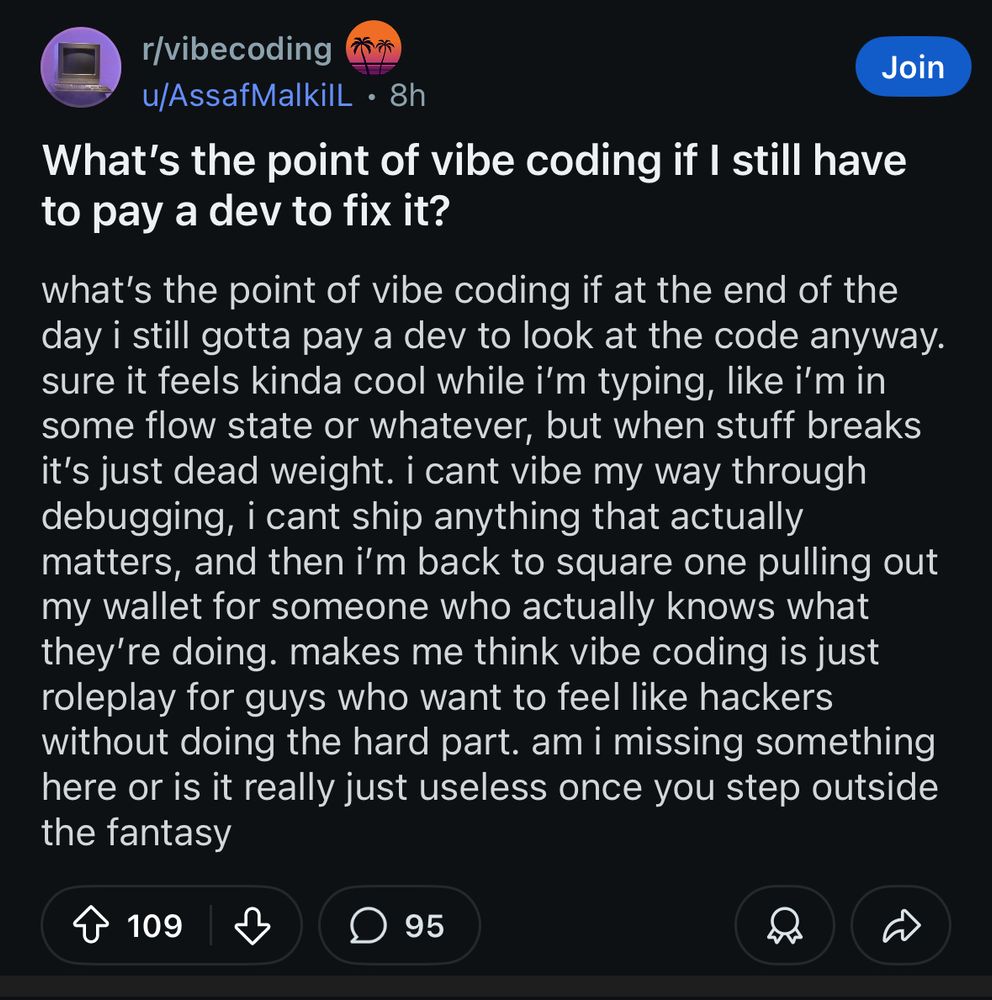
reddit post screenshot. what’s the point of vibe coding if i still have to pay a dev to fix it? what’s the point of vibe coding if at the end of the day i still gotta pay a dev to look at the code anyway. sure it feels kinda cool while i’m typing, like i’m in some flow state or whatever, but when stuff breaks it’s just dead weight. i cant vibe my way through debugging, i cant ship anything that actually matters, and then i’m back to square one pulling out my wallet for someone who actually knows what they’re doing. makes me think vibe coding is just roleplay for guys who want to feel like hackers without doing the hard part. am i missing something here or is it really just useless once you step outside the fantasy
you’re so close
19.08.2025 11:38 — 👍 3961 🔁 636 💬 113 📌 65
Why would anyone want to be a scientist?
It is difficult to fathom why anyone intelligent enough to be a scientist would actually choose to be one. Doing good science requires the utmost exertion of body, mind and spirit, yet is consistently...
"why [would] anyone intelligent enough to be a scientist choose to be one [given] the unfavorable risk-to-reward ratio "?
One of the most intelligent people you could meet offers some answers: having ideas, watching them develop, and sharing them journals.biologists.com/jcs/article/...
15.08.2025 16:16 — 👍 117 🔁 40 💬 5 📌 8

Why would anyone want to be a scientist? There are the pleasures of:
1. Having the initial idea or insight,
2. Watching the idea develop into new experiments or a new model, and
3. Telling others.
Martin A. Schwartz
Check out also his Night Science podcast episode: podcasts.apple.com/us/podcast/7...
15.08.2025 20:29 — 👍 63 🔁 17 💬 3 📌 0

Defending US democracy: the role of scientists
@science.org
www.science.org/doi/10.1126/...
14.08.2025 18:05 — 👍 297 🔁 124 💬 8 📌 5

FRAUD ALERT: I have nothing to do with these phony books using my name on the cover. I have not written a memoir or partnered/inspired any cookbooks!
Attempts to get Amazon to take these down have gone nowhere.
14.08.2025 22:11 — 👍 332 🔁 150 💬 19 📌 17
YouTube video by 3Blue1Brown
But how do AI videos actually work? | Guest video by @WelchLabsVideo
New video on the details of diffusion models: youtu.be/iv-5mZ_9CPY
Produced by Welch Labs, this is the first in a short series of 3b1b this summer. I enjoyed providing editorial feedback throughout the last several months, and couldn't be happier with the result.
25.07.2025 12:27 — 👍 146 🔁 13 💬 2 📌 3
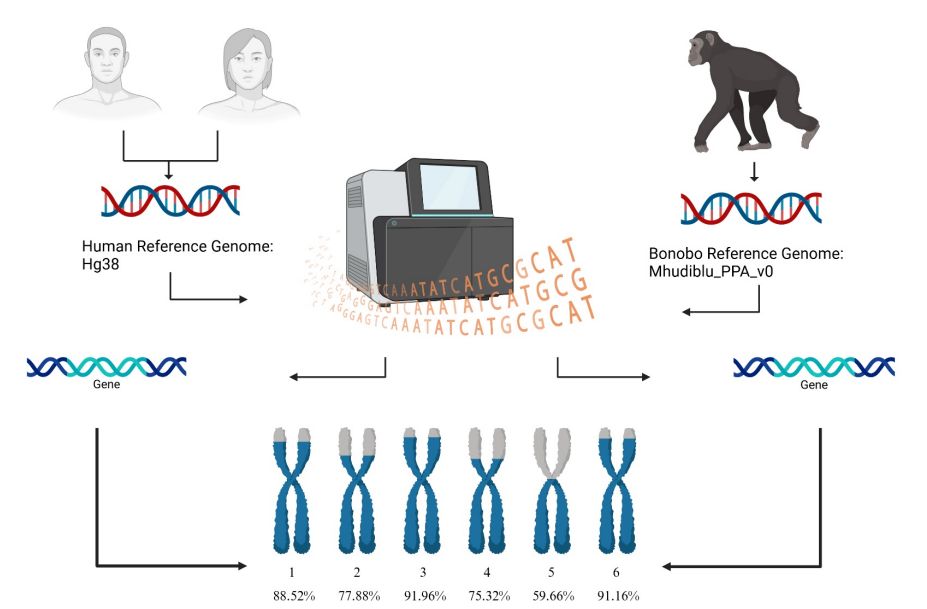
Unbiased whole genome comparison of Pan paniscus (bonobo) and Homo sapiens (human) through a novel sequence match-based approach. #WGS #GenomeComparison #Genomics #Bioinformatics @biorxiv-bioinfo.bsky.social 🧬 🖥️
www.biorxiv.org/content/10.1...
06.07.2025 17:05 — 👍 3 🔁 2 💬 0 📌 0
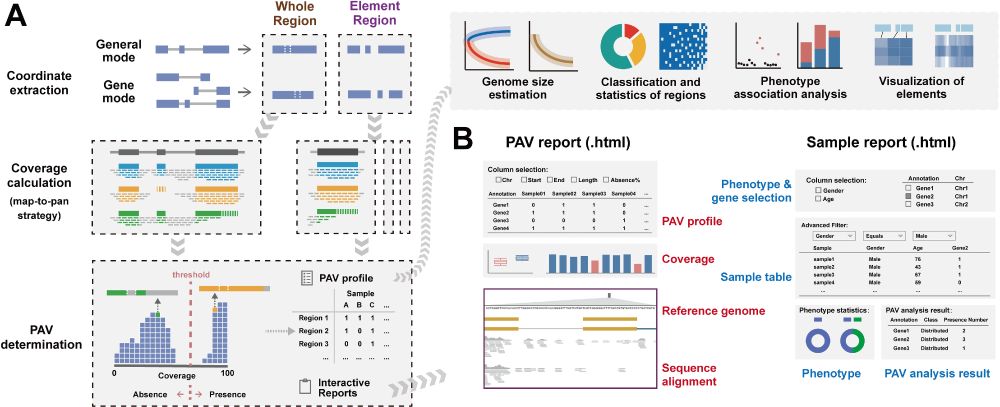
Fig 1. Overview of APAV pipeline.
(A) Workflow for PAV calling and consequent analysis. The coordinates of the target region are first extracted from a GFF file or BED file. The coverage of both the whole region and the element region is calculated based on the BAM files. The PAVs are determined according to their coverage, and interactive reports are generated. Subsequent analyses can be performed using the PAV tables, including genome size estimation, classification and statistical analysis, phenotypic association analysis, and visualization of elements. (B) Interactive analysis reports. The PAV report presents PAV tables, coverage data, pangenome sequences, genome annotation, and sequence alignments. The sample report presents sample tables, phenotype information, and real-time PAV analysis results.
https://doi.org/10.1371/journal.pcbi.1013288.g001
APAV: An advanced pangenome analysis and visualization toolkit
journals.plos.org/ploscompbiol...
13.07.2025 16:22 — 👍 4 🔁 2 💬 0 📌 0

Header text: "HEY YOU! YES YOU! TAKE THIS SHIT OUT OF LINKS!"
This is followed by a couple example urls, with a red square highlighting the sections with source identifiers. The rest of the text is as follows:
Source identifiers are used to track your activity on a site. Where you came from, what device you use, and even who you talk to. Whether it's written clearly in the url or tied to a random string of characters, it's assigned to your activity.
When you send a link containing a source identifier to somebody and they click it, it signals to the website that you two are connected. And that data goes right back to the website operators, and thus their advertisers.
Whenever you select "share" or "copy link" on a social app or website, it creates a link like this. If you give even the smallest shit about online privacy, it's important to remove them. Everything after the "?" symbol can be removed without issue, especially sections starting with "si=" or "utm_source="
I saw an infographic a couple years ago about how to remove source identifiers from links and why it's important, but I can't find it again and too many people I know are sending me links with them so here's an infographic straight from the oven
11.07.2025 22:22 — 👍 14846 🔁 8576 💬 208 📌 365
Caine-Hackman!!
13.07.2025 17:38 — 👍 38 🔁 9 💬 0 📌 0
Join the Bioconductor Developer Forum - R & Rust in Bioinformatics
🗓️ 28 July, 17:00–18:00 CEST
Discussing extendr, Rust packages, and real-world use cases.
🔗 blog.bioconductor.org/posts/2025-0...
#Bioconductor #RStats #RustLang #Bioinformatics
11.07.2025 12:46 — 👍 12 🔁 6 💬 0 📌 0

Indeed
20.06.2025 12:37 — 👍 652 🔁 122 💬 10 📌 1
Prof of Ed Psych and Learning Sciences | Making Tech Work For Us, Again | APA & AERA Fellow | Self-regulated learning, epistemic cognition, digital literacy | Journal and Handbook Editor | Book Author | Views are my own. https://linktr.ee/jeffgreene
Cutting-edge research, news, commentary, and visuals from the Science family of journals. https://www.science.org
A lab at the MRC LMB, Cambridge. Interested in dynein, microtubules, cytoskeleton, electron microscopy and much more.
Anthropologist - Bayesian modeling - science reform - cat and cooking content too - Director @ MPI for evolutionary anthropology https://www.eva.mpg.de/ecology/staff/richard-mcelreath/
Mathematician at UCLA. My primary social media account is https://mathstodon.xyz/@tao . I also have a blog at https://terrytao.wordpress.com/ and a home page at https://www.math.ucla.edu/~tao/
A programming language empowering everyone to build reliable and efficient software.
Website: https://rust-lang.org/
Blog: https://blog.rust-lang.org/
Mastodon: https://social.rust-lang.org/@rust
Alt-Account of @cripdyke.bsky.social
Tracking persons politically arrested, detained, or disappeared by the Trump regime since March 9, 2025.
Michele Banks
Artist inspired by science
Friend to squirrels
https://www.etsy.com/shop/artologica/
PhD. 🏳️🌈
Immunology, aging and bioinformatics enthusiast. @MSCActions postdoc at @i2sysbio.bsky.social @csic.es
Comics as thinking. Eisner winner. Unflattening from HarvardUP (http://bit.ly/1vENIO7). Former Detroiter/NYer/YYC/now assoc prof SFSU-Comics Studies!
https://spinweaveandcut.com/
Tags #comics
#Unflattening #Nostos
Designer, journalist, and professor.
Author of 'The Art of Insight' (2023) 'How Charts Lie' (2019), 'The Truthful Art' (2016), and 'The Functional Art' (2012). NEW PROJECT: https://openvisualizationacademy.org/
I speak ACTG 🧬 | PhD Candidate at #INCLIVA | Genomic bioinformatics in cardiovascular diseases | Collaborating in #I2SysBio and #ITI-UPV
Delivered effective, efficient, and secure digital services for the American people until we were forced to stop on March 1, 2025. Not an official government account. Reposts are not endorsements. Our new website: https://18f.org/ #AltGov
Mobilizing the fight for science and democracy, because Science is for everyone 🧪🌎
The hub for science activism!
Learn more ⬇️
http://linktr.ee/standupforscience
Account of the Saez-Rodriguez lab at EMBL-EBI and Heidelberg University. We integrate #omics data with mechanistic molecular knowledge into #opensource #ML methods
Website: https://saezlab.org/
GitHub: https://github.com/saezlab/
Finished just outside the top 3 Most Responsible Person in our 4 person household for a record 13 years.
Mostly I am just going to share cool sarcoma and epigenetics papers that I want to read 🧬🧪, with a sprinkling of the cutest dogs ever (mine🐶🐶) and maybe some food and 🏈!
We make free, open-source software for data scientists like the RStudio IDE.
We're formerly known as RStudio. You can always download our open-source IDE here. https://posit.co/download/rstudio-desktop/
Prof at #UCDavis
Interests: evolution, ecology, function & phylogenomics of host-microbe/microbiome symbioses; #openscience; #birds; baseball; T1D
Lab phylogenomics.me
Pics jonathaneisen.smugmug.com
Links linktr.ee/jonathaneisen
TED go.ted.com/6WPm
























