
Small angel X-ray scattering
19.06.2025 20:49 — 👍 60 🔁 8 💬 0 📌 0@grant.rotskoff.cc
Statistical mechanic working on generative models for biophysics and beyond. Assistant professor at Stanford. https://statmech.stanford.edu

Small angel X-ray scattering
19.06.2025 20:49 — 👍 60 🔁 8 💬 0 📌 0
Big fan of this perspective:
07.05.2025 18:46 — 👍 35 🔁 6 💬 2 📌 0The plan at FutureHouse has been to build scientific agents for discoveries. We’ve spent the last year researching the best way to make agents. We’ve made a ton of progress and now we’ve engineered them to be used at scale, by anyone. Free and on API.
01.05.2025 16:06 — 👍 13 🔁 3 💬 1 📌 2What an incredibly cool paper! While knot theory strictly applies to closed curves, Tommy, @smnlssn.bsky.social , and @paulrobustelli.bsky.social show that writhe, a knot "non-invariant" that changes with smooth deformations, provides a meaningful descriptor for flexible conformations.
01.05.2025 05:49 — 👍 7 🔁 2 💬 1 📌 0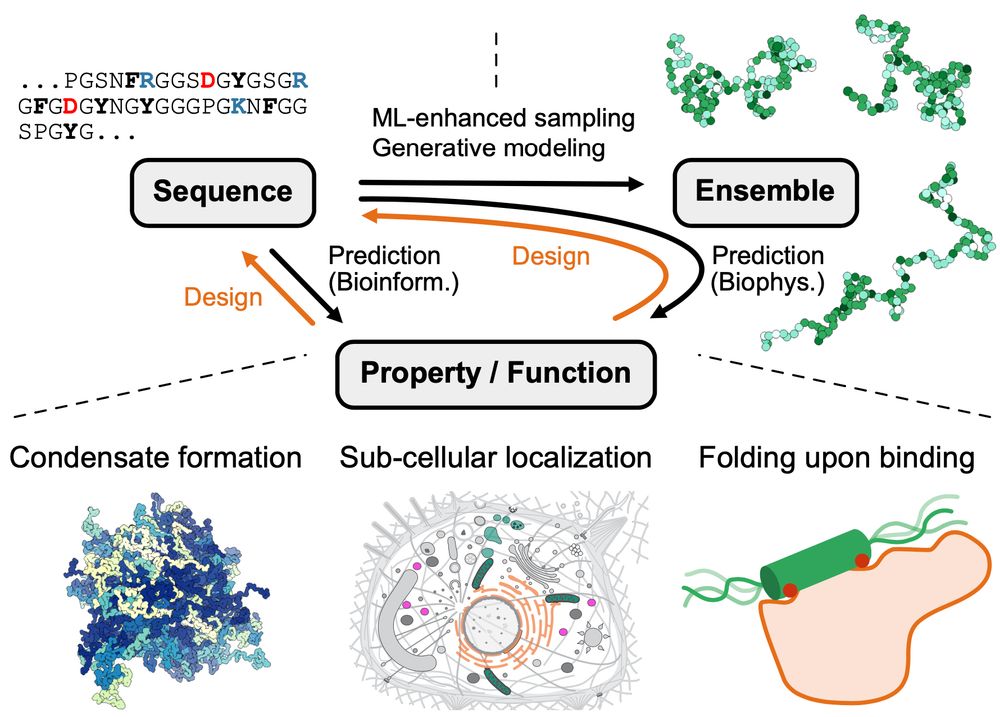
Figure from the paper illustrating sequence–ensemble–function relationships for disordered proteins. ML prediction (black) and design (orange) approaches are highlighted on the connecting arrows. Prediction of properties/functions from sequence (or vice versa, design) can include biophysics approaches via structural ensembles, or bioinformatics approaches via other hetero- geneous sources. The lower panels show examples of properties and functions of IDRs for predictions or design targets. ML, machine learning; IDRs, intrinsically disordered proteins and regions.
Our review on machine learning methods to study sequence–ensemble–function relationships in disordered proteins is now out in COSB
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei
Amazingly, this trick works. Due to improved algorithms for learning the score coming from the generative modeling, the applicability of this approach is very broad. I had spent several years making false starts on implementing the Malliavin calculus, but in the end, we found a route around it ;)
04.03.2025 18:45 — 👍 1 🔁 0 💬 0 📌 0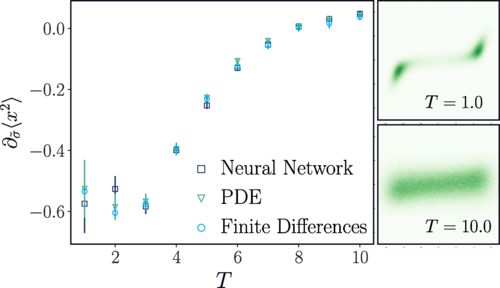
Jérémie Klinger found a simple trick back to Girsanov: you take the perturbation in the diffusion at the level of the Fokker-Planck equation and rewrite it to be included in the drift. The resulting drift then has a term proportional to \nabla \log \rho(x,t), what machine learners call the score!
04.03.2025 18:45 — 👍 1 🔁 0 💬 1 📌 0The “classical” strategy for diffusion sensitivities comes from financial mathematics and is called the Malliavin calculus, it’s very explicit for simple models like Black Scholes but for a general Langevin equation, it is no easy feat to compute the sensitivity.
04.03.2025 18:45 — 👍 0 🔁 0 💬 1 📌 0In many biological and active systems, diffusivity is highly spatially dependent, and the theory for perturbations in such cases is rather limited, largely based on beautiful work by Leticia Cugliandolo and work by Falasco and Baeisi, among many others.
04.03.2025 18:45 — 👍 0 🔁 0 💬 1 📌 0Far from equilibrium, it is not so easy: one needs to understand the dynamics, and this requires working with dynamical trajectories and their associated path measures. Classically, we do this using the Girsanov theorem, which constructs a “relative path measure” as we perturb the drift term.
04.03.2025 18:45 — 👍 0 🔁 0 💬 1 📌 0Computing response functions or “sensitives” requires understanding how an external perturbation drives the change in some observable. For equilibrium systems, Onsager taught us that this can be understood with correlation functions.
04.03.2025 18:45 — 👍 0 🔁 0 💬 1 📌 0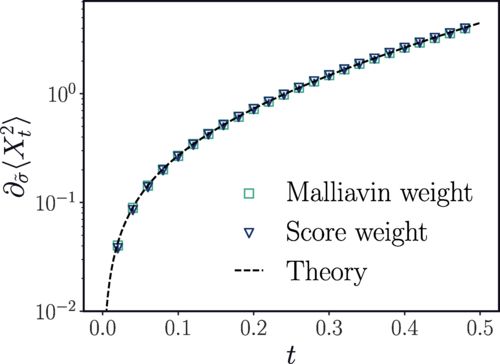
Excited to see our paper “Computing Nonequilibrium Responses with Score-Shifted Stochastic Differential Equations” in Physical Review Letters this morning as an Editor’s Suggestion! We uses ideas from generative modeling to unravel a rather technical problem. 🧵 journals.aps.org/prl/abstract...
04.03.2025 18:45 — 👍 10 🔁 2 💬 1 📌 0
Applications for the FutureHouse Independent Postdoctoral Fellowship are due in two weeks! $125k annual stipend, full access to our resources, be coadvised by world class professors and apply our AI science agents to make new discoveries. Apply!
Details here: www.futurehouse.org/fellowship
Ten simple rules for developing good reading habits during graduate school and beyond
To me, the most important are:
Read often, read broadly (incl. older papers and outside your field), and learn to read some papers in detail and others more superficially (and quickly)
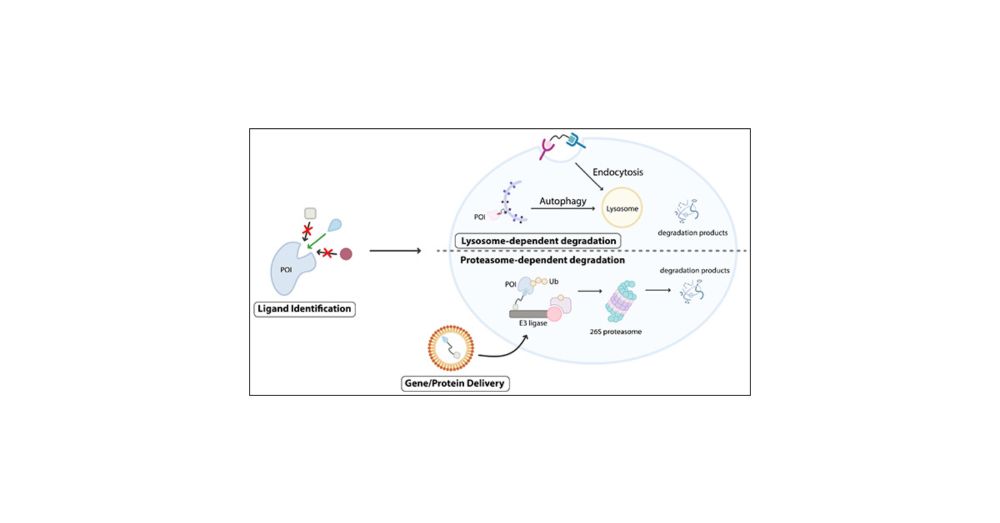
Excited to share this beast of a review on the potential of protein-based degraders from trainees who are all not on the Sky! Herein we cover choice of binders, selection strategies, E3 ligases, and DELIVERY! pubs.acs.org/doi/10.1021/...
18.01.2025 01:07 — 👍 40 🔁 10 💬 0 📌 1I am hiring a postdoctoral scholar with a start date summer or fall 2025. Projects will be focused on thermodynamically consistent generative models, broadly defined. If you’re interested, please send a CV and one paragraph about why you think you’d be a good fit to rotskoff@stanford.edu
23.12.2024 17:31 — 👍 48 🔁 22 💬 0 📌 0Lot of cool stuff in here. Consistent with my working hypothesis that the main scientific utility of LLMs at the moment is plain old NLP
22.12.2024 17:30 — 👍 9 🔁 1 💬 0 📌 0Really cool opportunity via futurehouse. Come work with them and collaborate with us at Stanford!
19.12.2024 17:55 — 👍 4 🔁 0 💬 0 📌 0
If you didn't see our poster at NeurIPS on how to make diffusion model inference fast, you can always read the paper here: arxiv.org/abs/2405.15986
13.12.2024 16:22 — 👍 10 🔁 0 💬 0 📌 0
also I must say often when I read new methods being pre-printed, while I appreciate the eagerness to make a splash, many folks seem unaware of the long history of this field & its assessments - to their detriment
If in CADD, pls read through D3R's last paper
pubmed.ncbi.nlm.nih.gov/31974851/
Can verify that the code works, too :)
07.12.2024 05:23 — 👍 0 🔁 0 💬 0 📌 0@franknoe.bsky.social presented this very impressive work at a fantastic @cecamevents.bsky.social workshop this week. I’m very excited to take a deep dive into the details this weekend!
06.12.2024 16:34 — 👍 15 🔁 2 💬 0 📌 0If you're at NeurIPS next week come see our spotlight poster led by Yinuo Ren and Haoxuan Chen! We use the parallel sampling technique to rigorously establish a big acceleration for diffusion model inference! neurips.cc/virtual/2024...
03.12.2024 21:55 — 👍 9 🔁 2 💬 0 📌 0
If you're talking about order in the dynamics itself, then dynamical large deviation theory is very relevant and the canonical review is by Hugo Touchette arxiv.org/abs/0804.0327
02.12.2024 22:38 — 👍 3 🔁 0 💬 1 📌 0There's no easy way to do this in general, but computing the stationary distribution for a nonequilibrium dynamics might be a possible in some low dimensional system or systems with special structure ( ones where you can represent the distribution with tensor networks). Simulations otherwise...
02.12.2024 22:38 — 👍 0 🔁 0 💬 1 📌 0Thanks for looping me in @erikhthiede.bsky.social If by order, you mean that there's some parameter that stabilizes to stationary or periodic steady state, then there the most general solution is simply solving for the stationary distribution or long time limit of the expectation of the order param
02.12.2024 22:38 — 👍 1 🔁 0 💬 1 📌 0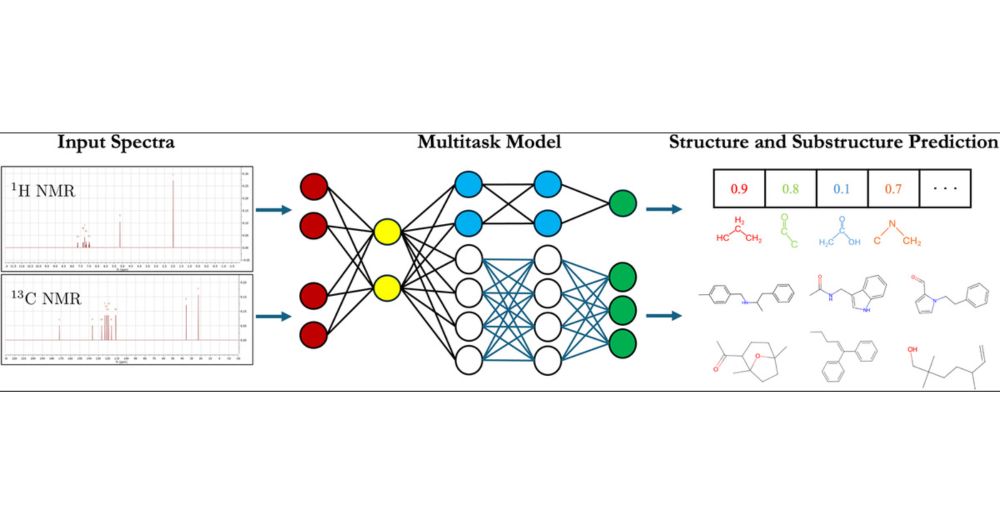
Chemists use NMR spectroscopy to identify molecules, but interpreting spectra is laborious and error prone. We show the process can be automated end-to-end using a well-designed Molecular GPT. Importantly, we also make predictions of substructures for interpretability. pubs.acs.org/doi/10.1021/...
13.11.2024 18:22 — 👍 12 🔁 3 💬 1 📌 0