In sum, we present the first de novo benchmark of protein-carbohydrate using AF3 and similar methods to identify the protein-sugar interactome at a structural level!
Once again, would like to thank my coauthors, who were integral to every step!
08.09.2025 17:12 — 👍 1 🔁 0 💬 0 📌 0
Although our work shows the impressive capabilities of these models; our work is limited in that we have few structures in our set, and those structures are mostly globular single-domain proteins bound to short carbohydrates - similar to our work on CAPSIF.
08.09.2025 17:10 — 👍 1 🔁 0 💬 1 📌 0

Previously, in PiCAP we uncovered the protein-sugar interactome, identifying over 7.5k human proteins that likely bind to carbohydrates. Since AF3 and other methods do well 85% of the time - can we use AF3 to predict these structures? Simple answer: No, not just yet on a high throughput scale
08.09.2025 17:05 — 👍 1 🔁 0 💬 1 📌 0

But, can these models tell us when they don't perform well - using their confidence metrics like ipTM or pLDDT? The simple answer - kinda. AF3 is incredibly overconfident, but there is a moderate trend in most models from prediction confidence and prediction accuracy.
08.09.2025 17:03 — 👍 1 🔁 0 💬 1 📌 0


Across the board, all models perform great at protein-carbohydrate docking, achieving 85% of top-5 structures with at least acceptable quality! However, the largest limitation is that overall performance decreases as saccharide length increases.
08.09.2025 16:59 — 👍 1 🔁 0 💬 1 📌 0

In this work, we developed a unique test set of 20 proteins to evaluate the performance of AF3, Boltz-1, Chai-1, DiffDock, and RFAA at protein-carbohydrate docking. We additionally made a new metric: DockQC, inspired by protein-protein docking to evaluate their performances across many test cases!
08.09.2025 16:57 — 👍 1 🔁 0 💬 1 📌 0

My latest manuscript is now on BioRxiv: Evaluation of De Novo Deep Learning Models on the Protein-Sugar Interactome. where we benchmark AF3 at protein-carbohydrate docking. This work was only possible with Dr. Lei Lu @takeshita-sho.bsky.social and @jeffreyjgray.bsky.social
doi.org/10.1101/2025...
08.09.2025 16:52 — 👍 11 🔁 1 💬 2 📌 1
I’d like to thank my PI Dr @jeffreyjgray.bsky.social
and co-mentor Dr. Ronald L Schnaar for their advise on this project. I’d also like to thank Sergey Lyskov for implementing the ROSIE server!
17.03.2025 19:05 — 👍 0 🔁 0 💬 0 📌 0
I would also like to thank the @rosettacommons.bsky.social at large, especially Yijie Luo for providing us with their designed carbohydrate non-binder dataset (which they used to train their CLIMBS algorithm, also just published on BioRxiv!).
17.03.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
If you’d like to use CAPSIF2 or PiCAP, you can do so freely at our github link: github.com/Graylab/picap or you can use our server on ROSIE: r2.graylab.jhu.edu/apps/ !
17.03.2025 18:00 — 👍 0 🔁 0 💬 0 📌 0

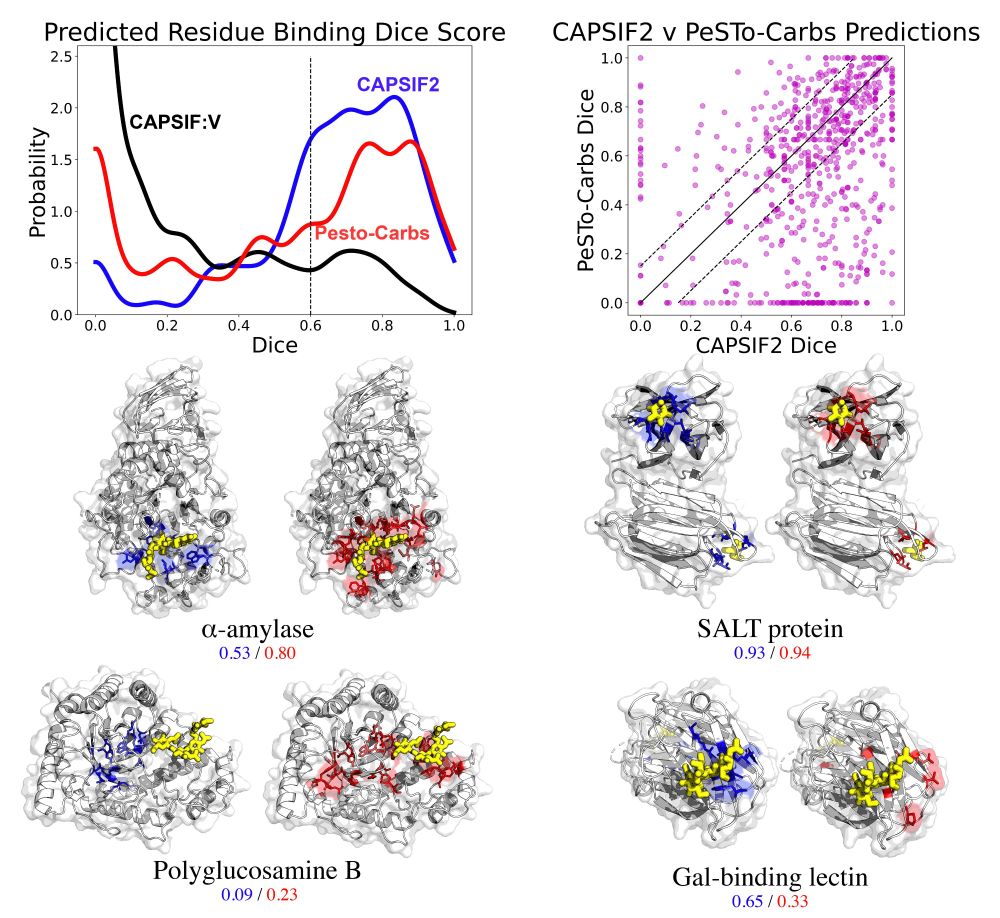
We also provide an updated model of our CAPSIF, a model for predicting carbohydrate binding regions of proteins. Comparing CAPSIF2 with @parthbibekar.bsky.social’s PeSTo-Carbs, finding their model outperforms CAPSIF2 on the TS90 dataset, but, CAPSIF2 modestly outperforms P-C on a larger dataset.
17.03.2025 17:59 — 👍 1 🔁 0 💬 1 📌 0
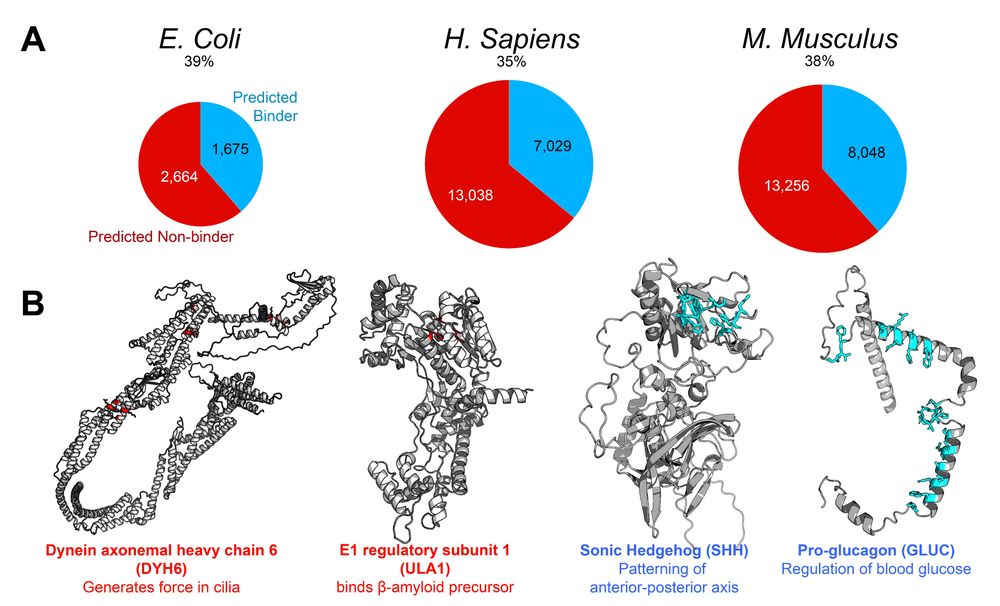
Now that the stage is set: what proteins bind to carbohydrates? We used PiCAP on three proteomes: E. Coli, mice, and humans and found that PiCAP predicts 35-40% of proteins bind to carbohydrates. And we provide a table of all proteins and their predictions for open scientific use and validation!
17.03.2025 17:56 — 👍 0 🔁 0 💬 1 📌 0
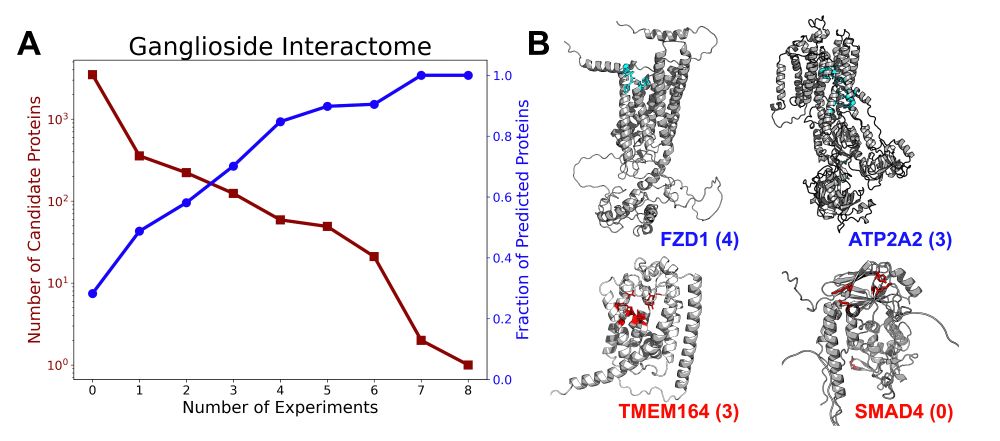
PiCAP does well on the test set -- but how does it do on other datasets that aren’t manually sculpted? We compare PiCAP to LectomeXplore finding a 99.5% agreement between the methods. We also interrogated the ganglioside interactome finding that PiCAP has a strong correlation with the experiments!
17.03.2025 17:54 — 👍 0 🔁 0 💬 1 📌 0
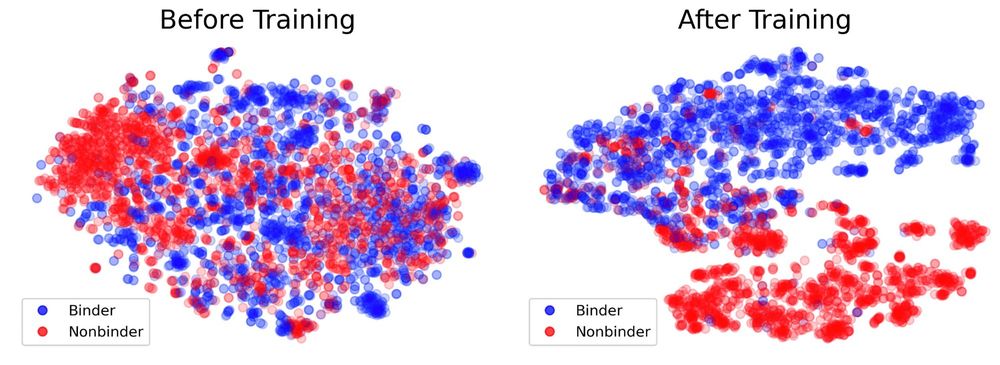
To do this we developed a deep learning (DL) model named Protein interaction of CArbohydrate Predictor (PiCAP). We find that PiCAP has approximately a 90% accuracy on identifying carbohydrate binding proteins, with limitations primarily on proteins that have reduced evolutionarily evidence (Abs).
17.03.2025 17:52 — 👍 0 🔁 0 💬 1 📌 0

It all started by asking just a simple question: “What proteins bind carbohydrates?” Lectins bind carbs, but, there are myriads of other proteins that bind carbohydrates (e.g. RTKs). And so we set out to find that out: can we find all the proteins in the human proteome that bind carbohydrates.
17.03.2025 17:51 — 👍 0 🔁 0 💬 1 📌 0
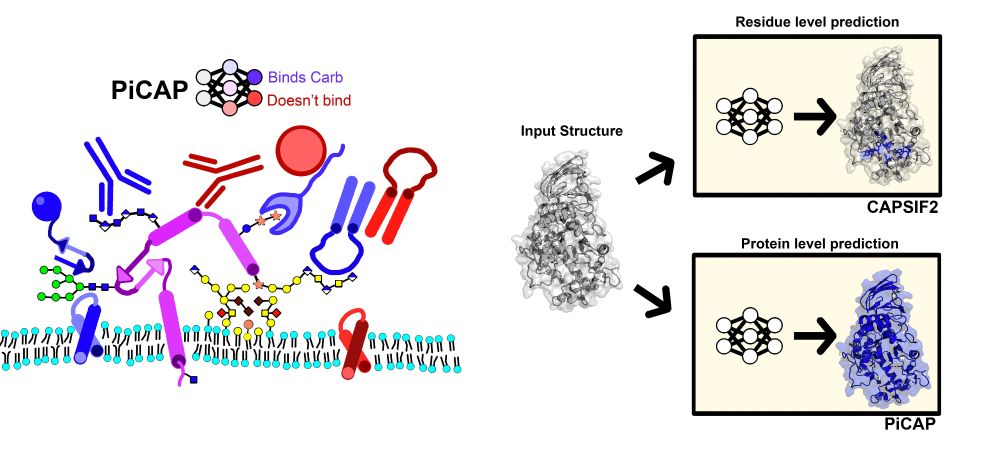
I’m happy to announce my latest paper has been released as a preprint on BioRxiv: Predictions from Deep Learning Propose Substantial Protein-Carbohydrate Interplay. This paper was only able to happen thanks to both @jeffreyjgray.bsky.social and Dr. Ronald L Schnaar
www.biorxiv.org/content/10.1...
17.03.2025 17:49 — 👍 2 🔁 0 💬 1 📌 1
a photo of people giving testimony in the House Judiciary Committee

a photo of Delegate Elizabeth Embry and advocates from Bike AAA, Bikemore, and lawyers who testified in favor of comparative negligence for vulnerable road users.
Today with sponsor Del. Elizabeth Embry we launched the fight to bring justice to vulnerable road users by replacing Maryland's contributory negligence standard with a fairer one that recognizes that a human being hit by a car driver suffers more than a driver in a crash.
20.02.2025 03:43 — 👍 14 🔁 4 💬 2 📌 0
Associate Professor of Biophysics at Johns Hopkins. Theory, modeling, and computation. Living systems and statistical mechanics. https://sites.krieger.jhu.edu/johnson-lab/
A PhD student in Biophysics @JHU. Interested in proteins and deep learning.
Senior Program Manager, Hopkins' 21st Century Cities Initiative.
Urban research with a Baltimore focus.
Ultrarunning.
At Bikemore, we believe a great city is a connected city. We focus on putting people before cars and expanding opportunity for all of Baltimore's residents.
Scientist, Assistant Professor at MIT biology, #FirstGen
PhD w/ Jeff Gray on protein docking/design @JHU ChemBE | Tweaking generative models for protein design @Generate Biomedicines | Runner | (chaotic) Protein geek (he/him/his)
Theoretical and computational biophysicist at Carnegie Mellon University. Loves lipid membranes, music, and art. (he/him) 🇩🇪🇺🇸
Johns Hopkins Chemical & Biomolecular Engineering Professor, Co-Director of RosettaCommons, protein engineer, Baltimore resident, father of two. he/him
ED of Bikemore. Cargo bike dad. Trying to make Baltimore City a little bit more friendly for walking, biking, and public transit. Occasional 🇨🇦 takes.
Physics of membranes and self-assembly || Biophysics postdoc at Johns Hopkins || Previously Penn State -> Carnegie Mellon || views are now yours, good luck || /dʒɪf/ ||
www.samuelfoley.com
PhD Candidate | EPFL
Structural bioinformatics
https://parthbibekar.github.io
Bioinformatics PhD @ UCSD | Research Fellow @ Ragon Institute at MGH, MIT, and Harvard | Founder
glycan metabolism, gene expression, NGS, pathology (he/him)
Boston MA, San Diego CA, Rochester NY, NYC NY
Biologist designing proteins
Postdoc at Fraunhofer IIP in Munich/Penzberg
official Bluesky account (check username👆)
Bugs, feature requests, feedback: support@bsky.app















