💸 Stem cell culture that’s ~95% cheaper and weekend-free: a win-win for PIs and trainees!
Our revised manuscript is online at ORE: open-research-europe.ec.europa.eu/articles/4-1...
Benchmarked across cell lines & differentiation protocols, including scRNA-seq — with lessons for #cultivatedmeat too!
20.09.2025 14:41 — 👍 23 🔁 8 💬 0 📌 0
Sad to hear that David Baltimore died. I am one of hundreds of scientists that David generously mentored, supported, and inspired. His influence on each of us was deep and lasting, and his impact on the world immeasurable. Arguably David Baltimore is one of the greatest scientists of any generation.
07.09.2025 21:50 — 👍 117 🔁 30 💬 5 📌 2

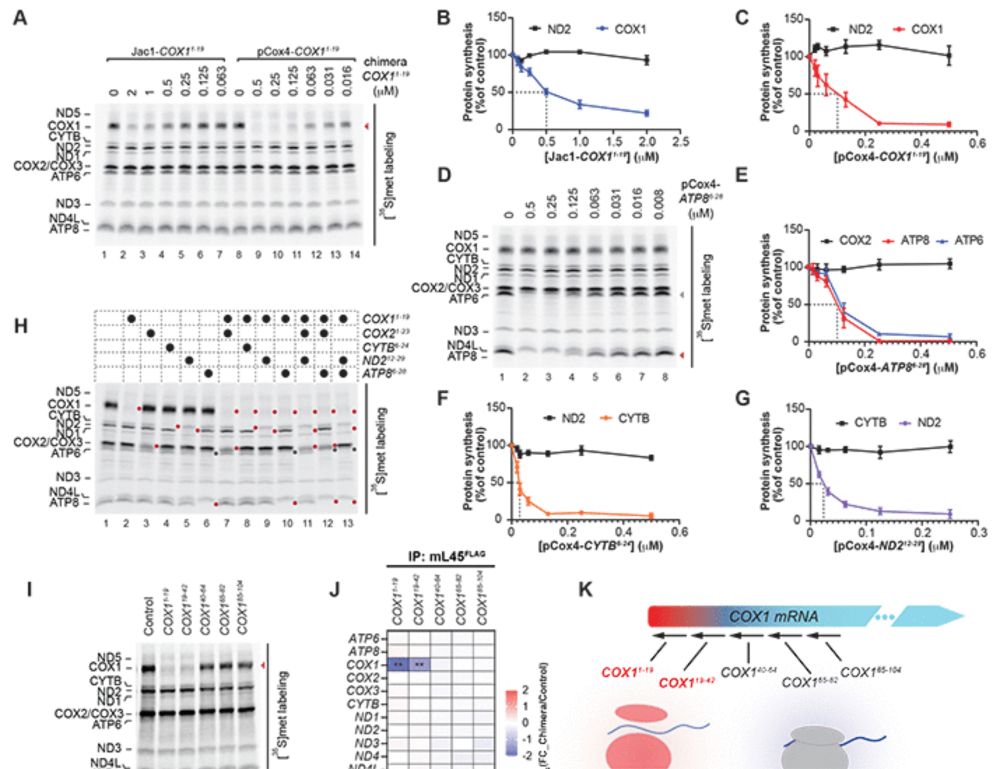
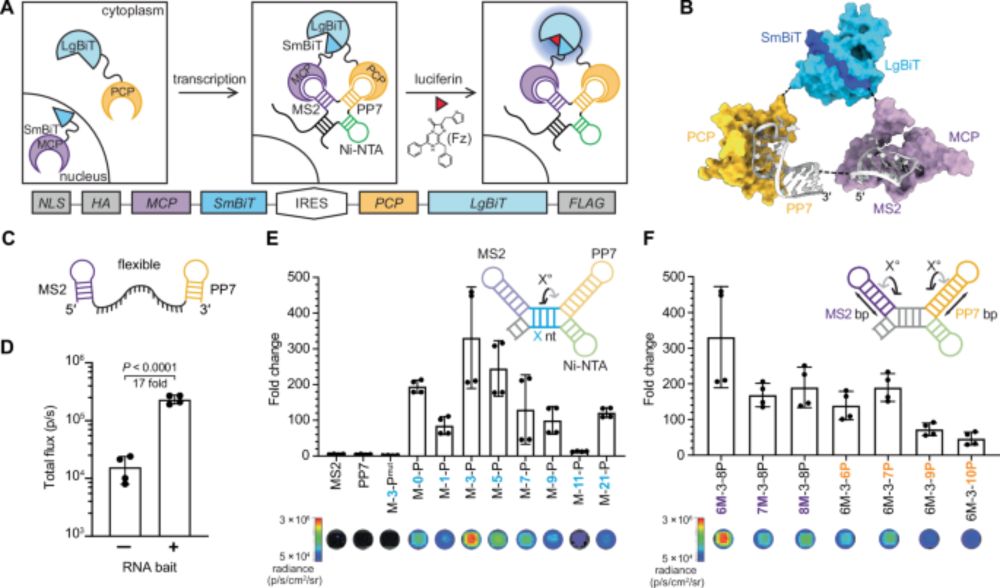
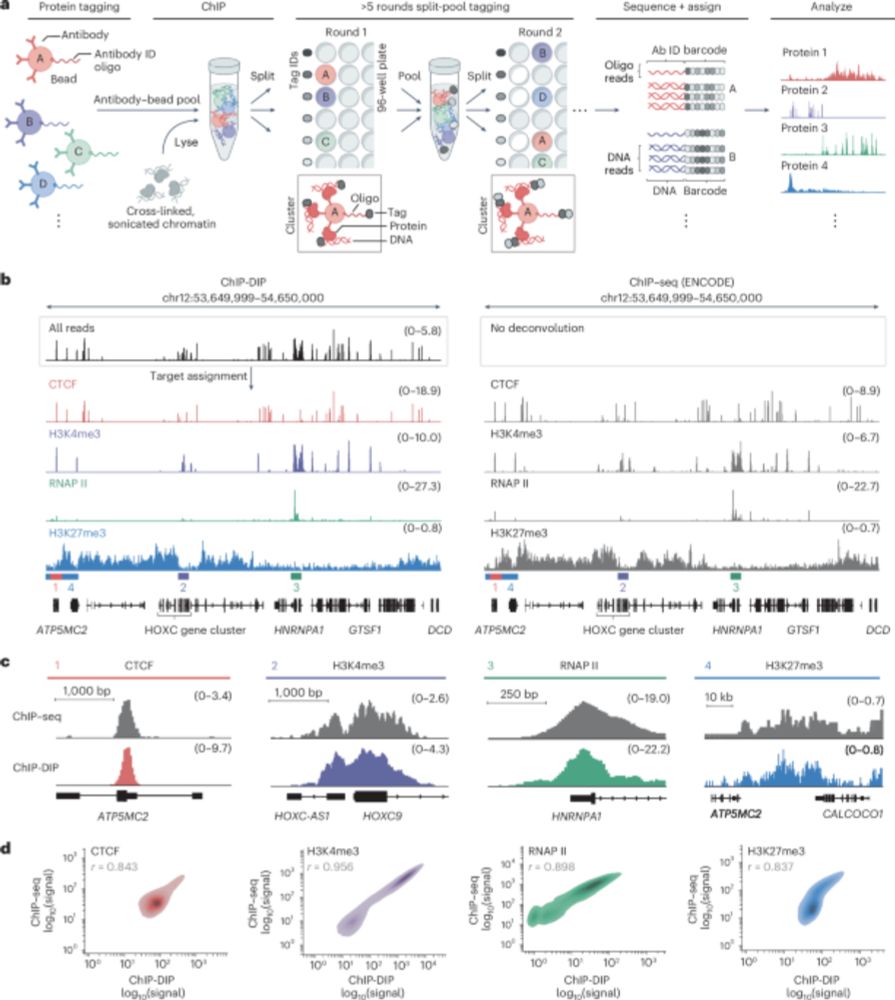
Many proteins bind RNA, yet we still don’t know what RNAs most bind because methods map one RBP at a time. In @cp-cell.bsky.social, with the Jovanovic lab, we describe SPIDR – a method for mapping the RNA binding sites of dozens of RBPs in a single experiment. www.sciencedirect.com/science/arti...
26.07.2025 19:15 — 👍 99 🔁 37 💬 1 📌 2
🚀 New from Cell: SPIDR is a game-changer—map dozens to hundreds of RNA-binding proteins simultaneously with single-nucleotide precision in one CLIP experiment. An exciting advance for RNA–protein interactomics! 🧬🔗https://doi.org/10.1016/j.cell.2025.06.042
27.07.2025 08:02 — 👍 8 🔁 2 💬 0 📌 0

I’m really pleased to share that our paper is out in Developmental Cell! NGN3 oscillates in the developing pancreas and controls the timing of cell differentiation via an incoherent feed-forward motif. For more details check out the comments below and DOI: 10.1016/j.devcel.2025.06.004
03.07.2025 13:10 — 👍 16 🔁 5 💬 3 📌 1

Our recent article on transposable elements and aging - which made the June cover - is the focus of a new Highlight.
"Highlight: 'Jumping Genes' Caught in an Aging Merry-Go-Round"
🔗 doi.org/10.1093/gbe/evaf094
🖌️ Miriam Merenciano
#genome #TEsky
11.06.2025 10:47 — 👍 21 🔁 9 💬 0 📌 1

Applications are now open! We are recruiting 20 Assistant Professors in a wide range of subject areas. We're looking for early-career researchers with strong scientific merits and future potential.
🔗 All positions: ki.se/en/about-ki/...
25.06.2025 10:00 — 👍 100 🔁 98 💬 0 📌 3
Our work on lineage tracing with somatic epimutations is finally out in @nature.com!
Check out the thread by @larsplus.bsky.social below.
EPI-clone is a revolutionary technique for lineage tracing and it will immediately impact how we trace clones everywhere.
Detailed protocols coming out soon!
21.05.2025 16:06 — 👍 52 🔁 13 💬 5 📌 0
New @nature.com
An ingenious way to track DNA replication patterns at the single-cell level that leads to cell heterogeneity and daughter cells that can potentially give rise to cancer
www.nature.com/articles/s41...
21.05.2025 17:17 — 👍 228 🔁 45 💬 3 📌 3
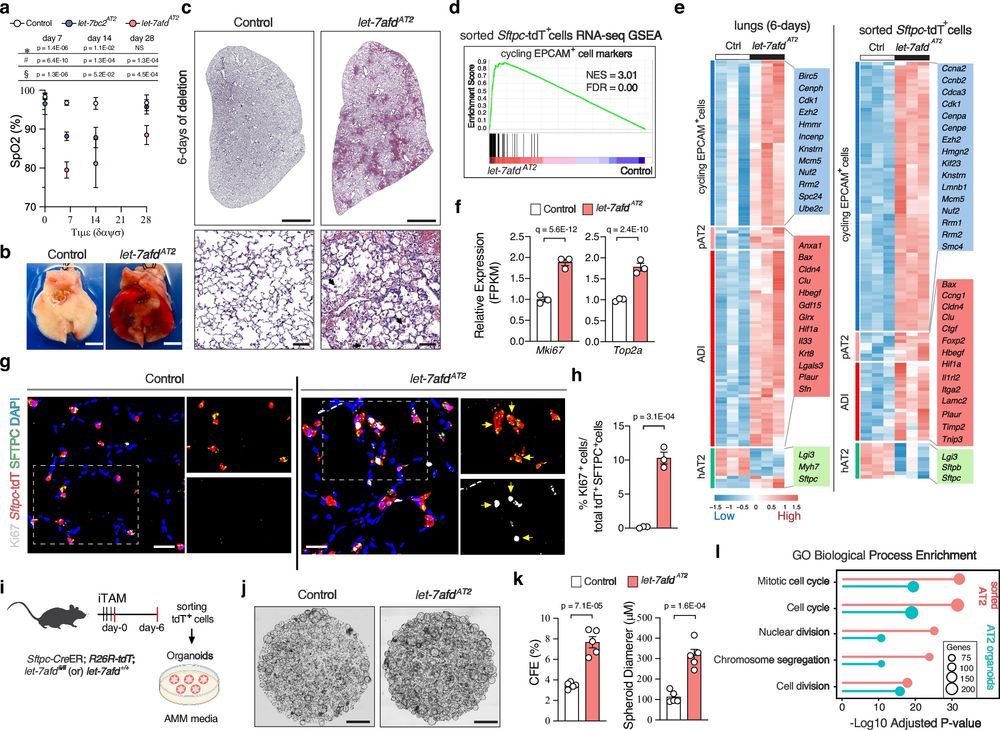
Congrats @micrornax.bsky.social on this study on let7 in lung injury/repair and thanks for including us as collaborators!
11.05.2025 20:30 — 👍 4 🔁 2 💬 0 📌 0
(4/n) Probing further with epigenomic experiments we demonstrated that Let-7 controls histone acetylation and methylation of H3K27 to reprogram the formation of profibrotic AT2 stem cells.
11.05.2025 01:41 — 👍 0 🔁 0 💬 0 📌 0
(3/n) We showed that Let-7 directly controls the expression of an extensive gene regulatory network comprised of transcription factors associated with fibrosis and cancer such as BACH1, EZH2, and MYC to safeguard the lung from scarring.
11.05.2025 01:37 — 👍 0 🔁 0 💬 0 📌 0
(2/n) We found that Let-7 serves as a molecular guardian of AT2 cells and prevents their abnormal growth and “transformation” into fibrotic scar forming KRT8+ transitional cells in pulmonary fibrosis.
11.05.2025 01:34 — 👍 0 🔁 0 💬 0 📌 0
Can’t wait for RNA2025! Chairing a new Education session… more details soon!
19.12.2024 00:13 — 👍 19 🔁 6 💬 1 📌 0
Our annual Dance Your Ph.D. contest is open for submissions until March 28 💃
The entry form is here (plus more details in the article linked below): www.science.org/content/page...
05.12.2024 19:07 — 👍 17 🔁 17 💬 1 📌 1
I'm really excited to see Prisca's work highlighted here @yegracechen.bsky.social!
05.12.2024 11:37 — 👍 4 🔁 2 💬 0 📌 0

Almost 1 month left to submit your abstract for the #Epigenetics2025 conference, organized by @gensocuk.bsky.social
We’ve put together a fantastic scientific program, and Northern Ireland is a wonderful destination to explore.
01.12.2024 12:30 — 👍 39 🔁 32 💬 0 📌 3
🚨 Update for #RNA science fans:
I checked on #Bsky registry, and here is a 'condensate' (😆) of all #RNA #RNAsky starter packs so far:
1. General RNA biology #1
go.bsky.app/RN6HnZp
21.11.2024 08:08 — 👍 134 🔁 56 💬 29 📌 4
Network of RNA labs across the greater Montreal area from McGill University, University of Montreal, UQAM, Concordia University, INRS, and industry. Holding monthly seminars.
JHU,PCCM, Outcomes After Critical Illness and Surgery (OACIS). Fellow @EmoryPCCM. Man united Fan🇸🇩🇺🇸🫁(tweets are my own)
Welcome to Random Shitty Thoughts
The weird, chaotic, slightly poetic mess that floats through my brain on a daily basis.
🔥 Blog by Dariusz Majgier. AI, fun facts, science & brilliant ideas:
👉 https://patreon.com/go4know
🔥 Get prompts, art styles & tutorials. Learn how to create Midjourney images/videos for FREE!
👉 Join me: https://patreon.com/ai_art_tutorials
Associate Professor in Molecular Genetics | University of Groningen | Chief R&D Officer @ Serna Bio | #RNA structural ensembles & dynamics | https://www.incaRNAtolab.com | Views are my own
Aging and cancer stem cell heterogeneity - ICREA research professor - Quantitative Stem Cell Dynamics lab
at IRB Barcelona 🇦🇷🇪🇺🇺🇸 fraticellilab.com
Follow @ats-assemblies.bsky.social for news and updates on the American Thoracic Society’s 14 Assemblies and Sections! 🫁
Molecular Cell Biologist focussing on Cancer and Endometriosis research.
Personal views.
lab at @UiTNorgesarktis studying microRNAs, paleotranscriptomics, biosystematics with a fable for flatworms.
MirGeneDB, MirMachine, MirMiner (soon!)
Data scientist & multiomic bioinformatics enthusiast, Assoc Professor at BCM, working on epigenetics, noncoding RNAs, infectious diseases, TB, IPF, and cancer
Anti-Smoker My Website campralquitsmoking.net See How I Quit 20 Years Ago Fought Nicotine 35 Years Supporting others who Quit 🚭 STL, Missouri ❤️To Promote Authors 📚 & Ozarks / Photographer 📷 @stegenmodem.bsky.social 💙
PhD candidate studying microRNA in C. elegans development. Music enthusiast.
Postdoc at UConn. RNA | RBP | 2OMe ~~~
Mobilizing the fight for science and democracy, because Science is for everyone 🧪🌎
The hub for science activism!
Learn more ⬇️
http://linktr.ee/standupforscience
20+ years in healthcare industry, sharing info in support of Healthcare Access for all. Former NIHer. Reposts intended to share info & don't = an endorsement. Views in notes are my own. 🛟
https://www.freepik.com/icon/health_8869931" Icon by Dragon Icons
Facts & strategy, in an authoritarian takeover.
Rightwing billionaires want to privatize NIH and use it to control universities.
We work to cure diseases like cancer.
Pers views. #science #medicine
Public health experts resisting harmful policies, fighting disinformation, and defending public health from within.
Not official CDC—but close enough for transmission.
Proudly #AltGov
Tips? signal altCDC.99, email altCDCtips@proton.me or DM
Promoting health and science and resistance against pseudoscience, disinformation, and fascism.
OG AltGov. that’s now here. ANTIFA!