🫠
29.01.2026 00:11 — 👍 0 🔁 0 💬 1 📌 0🫠
29.01.2026 00:11 — 👍 0 🔁 0 💬 1 📌 0
Excited to share our latest preprint: "SWARM: A Single-Molecule Workflow for High-Precision Profiling of RNA Modifications" www.biorxiv.org/content/10.6...
Led by Stefan Prodic #RNA #Epitranscriptomics #Nanopore 1/6

Also, @bonson-wong.bsky.social did a slow5curl release github.com/BonsonW/slow... to support these ex-zd compressed BLOW5
28.01.2026 23:50 — 👍 1 🔁 0 💬 0 📌 0
slow5tools v1.4.0 released github.com/hasindu2008/...
Many bit profiles for ex-zd lossy compression added by @hiruna72, who reduced 275TB of historical @nanopore rawdata at @GenTechGp to 172TB.
guide to lossy archive: hasindu2008.github.io/slow5tools/a...
paper: doi.org/10.1101/gr.2...
Now that the @nci-australia.bsky.social Gadi supercomputer in Australia has H200 GPUs, @nanopore basecalling is much faster than before. For those who are interested, I have updated my example scripts [https://github.com/hasindu2008/nci-scripts] to demonstrate how these can be used.
22.01.2026 05:20 — 👍 2 🔁 1 💬 0 📌 0congratulations!!!! 🎉
20.01.2026 06:17 — 👍 1 🔁 0 💬 0 📌 0
slorado v0.4.0-beta is released: github.com/BonsonW/slor...
- Support for RNA basecalling
- 20-35% performance improvement for super-accuracy basecalling
Work by @bonson-wong.bsky.social.
Yes that is the process.
Very detailed step are here:
hasindu2008.github.io/cornetto/doc...
If you run into obstacles when setup, please open an issue - will be helful for us to streamline the setup steps where possible.
Yes, it works with MinION.
I have detailed the steps here: hasindu2008.github.io/cornetto/doc...
Not sure if this is simple enough though :D
I made a small wrapper library for reading slow5/blow5 files in #julialang based on the C slow5lib. If anyone working with #nanopore sequencing is interested, you can find it here: https://codeberg.org/mzdravkov/Slow5.jl
05.12.2025 09:38 — 👍 10 🔁 7 💬 6 📌 0
Cornetto v0.2.0 is now released for using programmable selective nanopore sequencing for genome assembly.
GitHub: github.com/hasindu2008/...
Paper: nature.com/articles/s41...
Datasets: hasindu2008.github.io/cornetto/doc...
... and a banner made by Ira Deveson referring to ‘no boring bits’.

Congratulations Hiruna Samarakoon (yet to be on bluesky) for winning the #abacbs2025 “Torsten Seemann” Outstanding Bioinformatics Software Developer Award!!! 🎉🎉🎉
26.11.2025 05:01 — 👍 19 🔁 7 💬 0 📌 0Cornetto uses readfish by @minomatt.bsky.social for adaptive sampling & hifiasm by Hayou, @lh3lh3.bsky.social et al for assembly.
26.11.2025 04:12 — 👍 1 🔁 0 💬 0 📌 0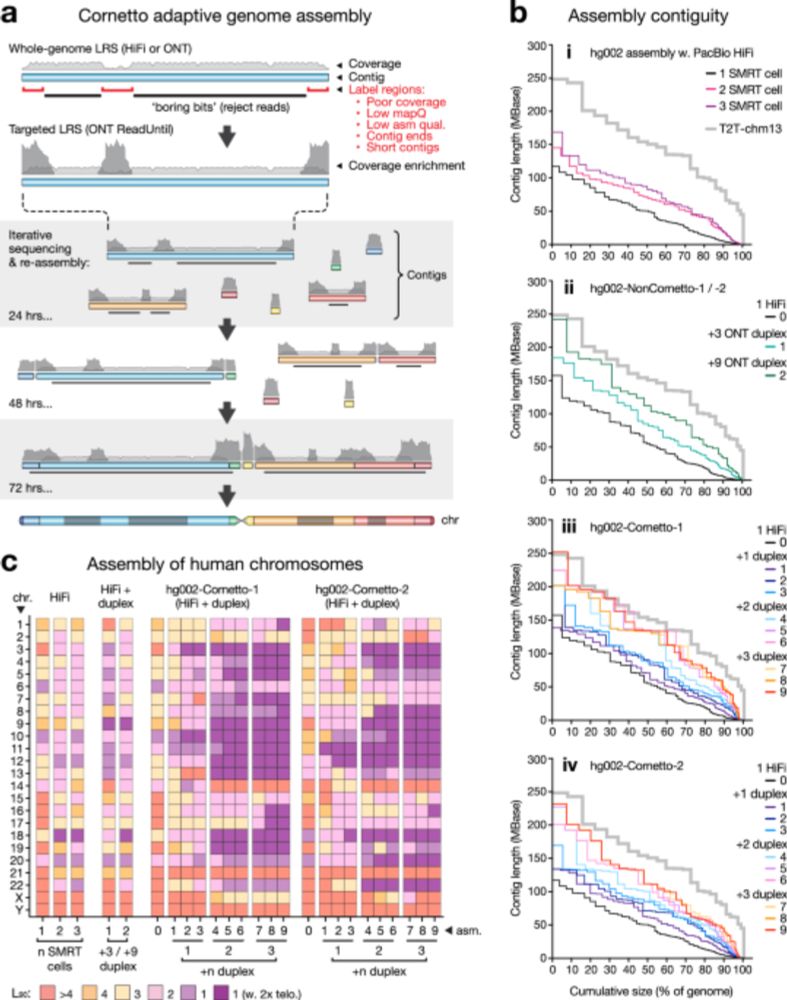
Our cornetto work is now published at www.nature.com/articles/s41...
It can do near-T2T assembly using @nanoporetech.com adaptive sampling
- with less 💸
- reference agnostic, so works for non-humans
- not just blood, even saliva
Just presented at #abacbs2025 yesterday.
With Slorado, now you have more choices for GPUs when basecalling @nanoporetech.com sequencing data. This is a work we collaborated on with AMD, led by
PhD candidate @bonson-wong.bsky.social (poster at #abacbs2025) and great to see being highlighted in the AMD blog: www.amd.com/en/blogs/202...

Thanks mate
Off the press just couple of hours before the talk
www.nature.com/articles/s41...
😂
25.11.2025 07:04 — 👍 0 🔁 0 💬 0 📌 0
possibly because ONT use their POD5 format which is not great on HDD. They could have adopted our slow5 format which we showed long ago that works for even HDD, before pod5 came in.
Now the pod5 (while indeed better than fast5) is still behind slow5 in this regards
academic.oup.com/gigascience/...
Sometime ago I changed a configuration file so this location is changed to our /data location permanently. Don't know if they have changed this option now.
30.10.2025 04:55 — 👍 0 🔁 0 💬 0 📌 0Thanks mate
21.10.2025 04:07 — 👍 0 🔁 0 💬 0 📌 0Thanks mate.
21.10.2025 04:06 — 👍 1 🔁 0 💬 1 📌 0Yeh, many better ways exist. We just showed how the most naive method can still keep up truck load volumes of data.
21.10.2025 04:05 — 👍 1 🔁 1 💬 0 📌 0In my personal opinion, more popular it is, more care should be taken to avoid compatibility problems. Adhoc implementation-driven changes can be avoided if enough thought is given at the design phase.
21.10.2025 04:04 — 👍 0 🔁 0 💬 0 📌 0
Just checked my logs and yes it is true.
The output produced was wrong - A simple program that sums up the signal values.
I just checked release logs of pod5 - 0.0.20 "Fix bug reading data via C API". So it could be a bug that got introduced in before.
@psy-fer.bsky.social @gigascience.bsky.social
16.10.2025 22:21 — 👍 0 🔁 0 💬 0 📌 0
Benchmark comparing SLOW5 and POD5 for nanopore raw signal data has now been published at
@GigaScience
academic.oup.com/gigascience/....
Some plots required a log scale - RAM usage and random access time.
For anyone interested, we uploaded another new HG002 @nanopore R10.4.1 (e8.2.1 enzyme) dataset with raw signals and super-accurate basecalls.
Reads lengths: 9.0 kbases median, 17.2 kbases mean
See:
gentechgp.github.io/gtgseq/docs/...

f5c v1.6 released. Now it supports a wide range of AMD GPUs. Something me and @bonson-wong.bsky.social worked on during a hackothon organised by CSC and @pawseycentre.bsky.social
github.com/hasindu2008/...
If you have any AMD GPUs lying around, help us test the binaries.

Cornetto v0.2.0-beta released
github.com/hasindu2008/...
Cornetto is not only an iterative assembly method for @nanoporetech.com, but also features some basic assembly evaluation tools that we implemented.
Suggestions and feedback welcome!

Meanwhile, questions by me and @psy-fer.bsky.social on POD5 writing opened months ago are yet to be answered by ONT.
It is very interesting that they have skipped those questions 🤣