The metagenomic datasets used in this study are from SPIRE. academic.oup.com/nar/article/...
If you are interested in the planetary microbiome, see also recent MicrobeAtlas preprint from von Mering group: www.biorxiv.org/content/10.1...
@chanyeong-kim.bsky.social
PhD | Microbiome researcher | Postdoc @Bork Group | EMBL-Heidelberg

The metagenomic datasets used in this study are from SPIRE. academic.oup.com/nar/article/...
If you are interested in the planetary microbiome, see also recent MicrobeAtlas preprint from von Mering group: www.biorxiv.org/content/10.1...
Generalists emerged as vehicles facilitating gene flow across ecologically disparate habitats, exemplified by the generalist-mediated HGT of an resistance island between the human gut and wastewater, with further dispersal to other habitats. This highlights human impact on the planetary microbiome.
21.07.2025 11:56 — 👍 1 🔁 0 💬 1 📌 0The framework enabled us to assess the gradient of prokaryotic generalism, from specialists that have adapted to and thrive in specific habitats to generalists capable of tolerating a broad range of environmental conditions.
21.07.2025 11:56 — 👍 0 🔁 0 💬 1 📌 0We delineated 40 microbiome habitat clusters, each representing distinct environmental contexts. Regardless of biogeography, microbiomes were primarily structured by host-associated or environmental conditions, with finer-scale structuring by factors such as host lifestyle and ocean temperature.
21.07.2025 11:56 — 👍 1 🔁 0 💬 1 📌 0
Our new preprint is out!
www.biorxiv.org/content/10.1...
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social
Our study emphasizes the importance of using standardized taxonomy to uncover features hidden within microbial dark matter. Our findings also offer significant translational potential for developing microbial biomarkers and therapeutic adjuvants for ICB treatment.
Thanks to all involved! :)
We discovered a pilus gene complex exclusive to TANB77. We showed that the synthesized pilin stimulates DCs through the TLR4 pathway. Administering pilin as an adjuvant to therapy in mice enhances ICB efficacy, with single-cell analysis revealing changes in the tumor immune microenvironment.
28.12.2024 13:56 — 👍 0 🔁 0 💬 1 📌 0We validated our findings through mouse experiments, showing that mice with higher gut TANB77 abundance, either naturally or through FMT, exhibited an improved response to anti-PD-1 therapy.
28.12.2024 13:56 — 👍 0 🔁 0 💬 1 📌 0Despite its prevalence as a human commensal, TANB77 remains an uncultured and understudied clade, unrecognized due to the limitations of conventional taxonomy. In NCBI taxonomy, TANB77 is confounded with unrelated taxa. We therefore refined our taxonomic classification by adopting GTDB taxonomy.
28.12.2024 13:56 — 👍 0 🔁 0 💬 1 📌 0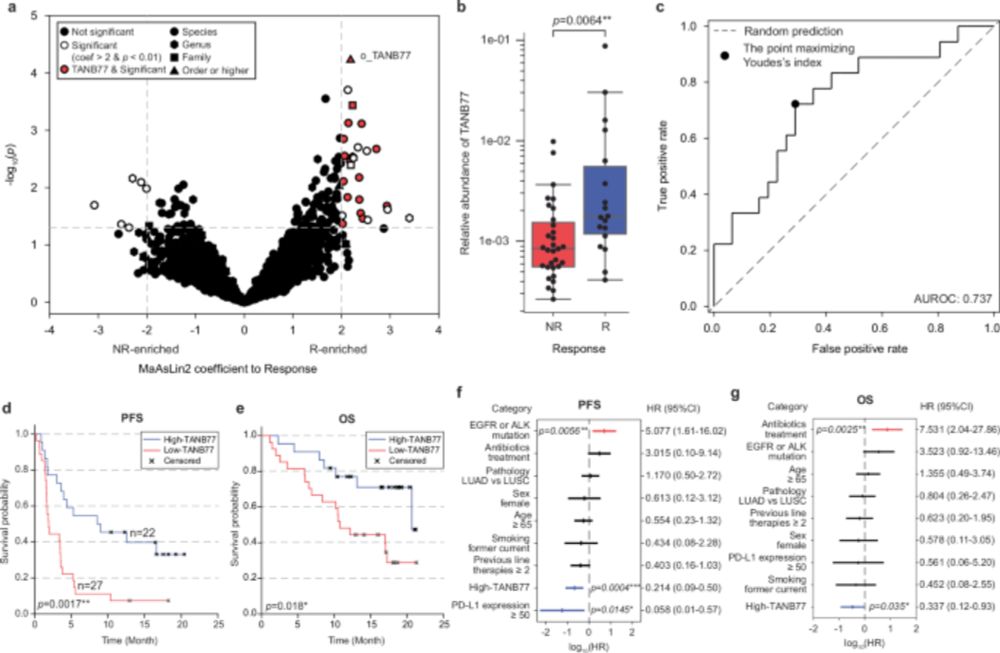
I’m excited to share our new paper published in Nature Communications!
In our work, we discovered TANB77, a bacterial clade previously obscured by a polyphyletic grouping in conventional taxonomy, as a reliable biomarker across diverse immunotherapy recipient groups.
www.nature.com/articles/s41...