Did you ever wonder what insights hide in the multiplets that are usually discarded in flow cytometry? 🫧 Find out in the first publication of my PhD, now out in Nature Methods! www.nature.com/articles/s41... (1/4)
07.08.2025 20:48 — 👍 33 🔁 17 💬 3 📌 0
proMGE
At #ISMBECCB2025, @agrekova.bsky.social will present her recent work on detecting mobile genetic elements in both reference and metagenome-assembled genomes in the Microbiome track on Thursday at 3 pm. Come for a sneak peek of the content in promge.embl.de version 2 (to be released in ~1 month)!
22.07.2025 07:08 — 👍 6 🔁 3 💬 0 📌 0
The MicrobeAtlas database: Global trends and insights into Earth’s microbial ecosystems www.biorxiv.org/content/10.1... #jcampubs
21.07.2025 16:45 — 👍 11 🔁 5 💬 0 📌 0
New preprint from the lab: "Planetary microbiome structure and generalist-driven gene flow across disparate habitats" analysing 85k metagenomes across the world, e.g. looking into generalism across habitats www.biorxiv.org/content/10.1... – see the thread below for more details!
#MicroSky 🖥️🧬🦠
21.07.2025 13:57 — 👍 23 🔁 12 💬 0 📌 0
Excited to share this @borklab.bsky.social preprint that I co-led with @chanyeong-kim.bsky.social and @podlesny.bsky.social. Thanks also to everyone else involved!
#Microbiology #Metagenomics #HGT #ARG
21.07.2025 12:01 — 👍 7 🔁 0 💬 0 📌 0
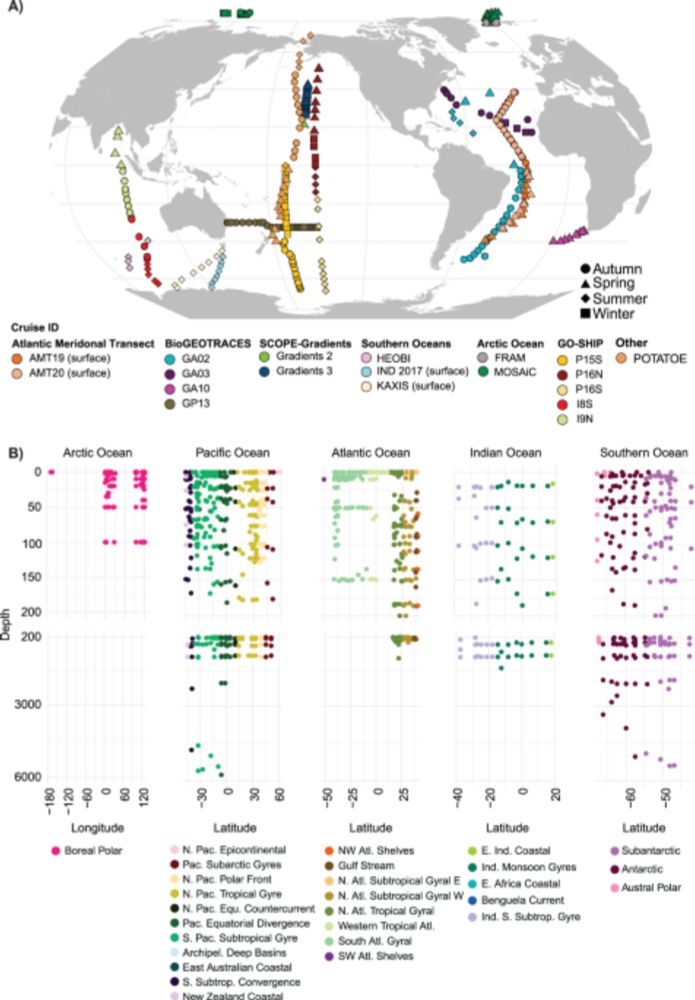
Characterizing organisms from three domains of life with universal primers from throughout the global ocean - Scientific Data
Scientific Data - Characterizing organisms from three domains of life with universal primers from throughout the global ocean
Please Re-Post. Our global GRUMP microbial ocean database is out in Scientific Data! Unfractionated, single universal PCR, with Archaea, Bacteria, and Eukaryotes all on the same scale with the same denominator. Pole to pole, depths to 6000m. Lots of metadata. www.nature.com/articles/s41...
02.07.2025 16:19 — 👍 46 🔁 31 💬 2 📌 2
The team's first preprint is out!
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
27.06.2025 20:09 — 👍 46 🔁 30 💬 1 📌 2
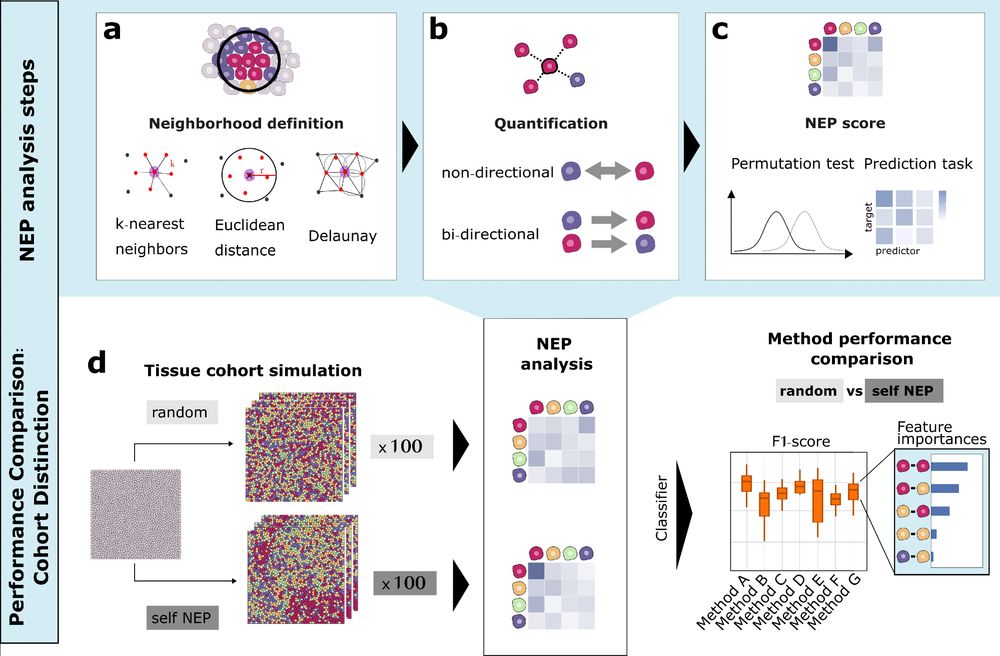
Schematic overview of NEP analysis steps (Neighborhood definition, Quantification and NEP score) and the systematic method performance comparison using simulated data for cohort distinction.
1/
Ever wondered how to best quantify cell-cell neighbor preferences in tissues?
We compared 9+ neighbor preference (NEP) methods for analysing spatial omics data and propose a novel approach that combines the most relevant analysis features which we call COZI 🔬✨
Read more: doi.org/10.1101/2025...
15.04.2025 15:50 — 👍 34 🔁 12 💬 1 📌 2
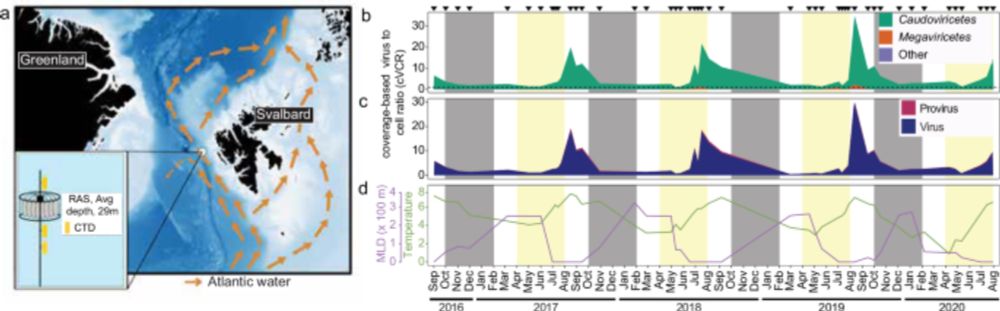
Our new study in @naturecomms.bsky.social showing the recurrent assembly of microbial modules over polar day and night, based on four years of autonomous sampling in the Arctic www.nature.com/articles/s41...
Fantastic teamwork by #AWI @mpimarinemicrobio.bsky.social @hhu.bsky.social & many others 🙂
04.02.2025 17:01 — 👍 11 🔁 8 💬 0 📌 0
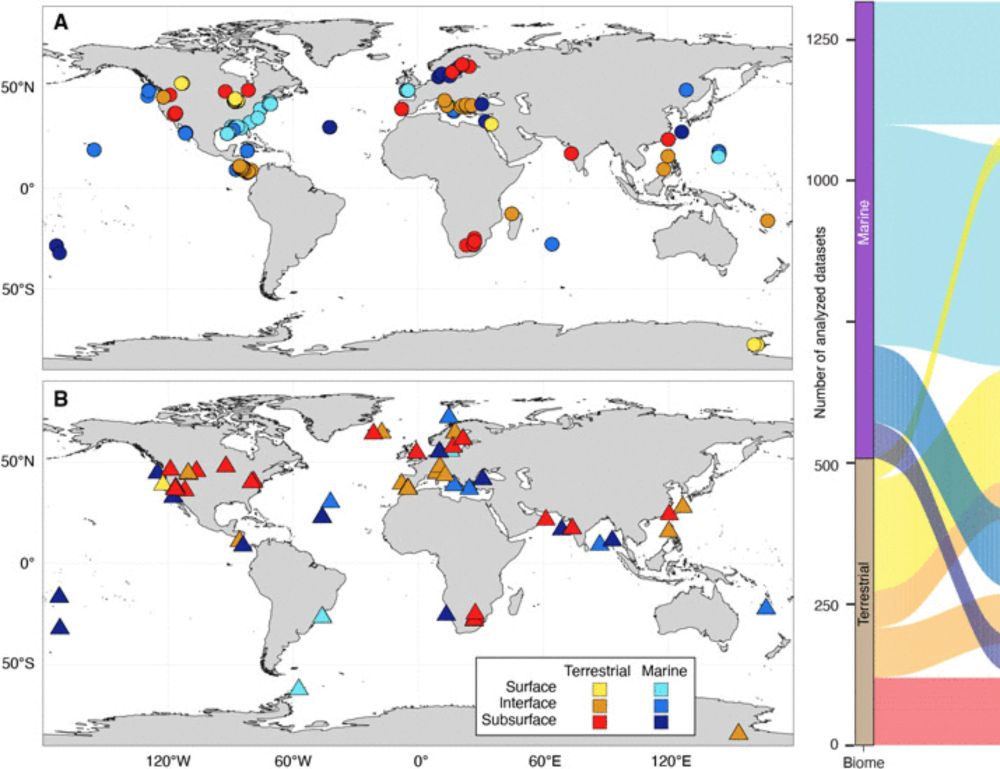
A global comparison of surface and subsurface microbiomes reveals large-scale biodiversity gradients, and a marine-terrestrial divide
The microbiomes of Earth’s surface and subsurface environments are different in composition, but similar in diversity.
Deep life is astonishing and diverse, and we have so much more to learn!
So happy that this study is out. Big thanks to co-first author @bellahda.bsky.social and all the collaborators!
www.science.org/doi/10.1126/...
@science.org @mblscience.bsky.social @simonsfoundation.org #deeplife #microbiome
18.12.2024 23:12 — 👍 84 🔁 38 💬 4 📌 7
Physician Scientist @UniHeidelberg.bsky.social. Love my family, science, medicine, traveling. Miss my friends around the world. Views are my own.
PhD student at EMBL Heidelberg
Interested in computational biology, biological image analysis and AI in medicine
Developing fast and easy methods for #phylogenetics and #bioinformatics | PhD in Bioinformatics | Postdoc @ Comparative Genomics Lab, UNIL/SIB🇨🇭| Formerly @ Steinegger Lab, SNU🇰🇷 | he/him
Group leader @EMBL Heidelberg | Archaea | Chromatin | Cryo-EM and cryo-ET | Evolution | Structure
Biomedical policy and climate-and-health reporter at Science. Author of The Vaccine Race. Book lover. History lover. Dog lover. Dual U.S.-Canadian citizen, proud Vancouver native. Signal me at: meredithwadman.13
Molecular microbiology (Corynebacterium, Listeria), live biosensors and biotechnology of bacteriocins and other fancy peptides @ Ulm University
Professor for Microbiology at Research Alliance Ruhr, Uni-DUE, #ERCSyg awardee, member of CRC RESIST
#microbiome #omics #microscopy #ecophysiology
Free time: Dad, weight training, cs2, nature, bbq, board games, pottery
🇵🇹 Staff Scientist @probstlab.bsky.social
(@unidue.bsky.social - 🇩🇪). Microbial genomics in the One Health context, biogeochemistry of cave microbiomes, alga-microbe symbioses.
Group for Environmental Metagenomics, led by @alexjprobst.bsky.social
Microbial, viral, & geochemical interactions by linking meta'omics & microscopy
Managed by @geomicrosoares.bsky.social
Microbiology Society: A world in which the science of #microbiology provides maximum benefit to society | microbiologysociety.org
lnk.bio/microbiosoc
Posts by Shankar Iyer, editor of Trends in Microbiology.
Postdoc fellow at EMBL (DE)
"views are my own"☘️🌊🦠🧪🌈
PhD student at UNC Charlotte l Marine microbial ecologist l Study giant viruses 🦠
🎓 Ghent University / Sorbonne alum
He/him
https://sites.google.com/uncc.edu/microbial-ecology-lab/home
Assistant Professor, University of North Carolina at Charlotte
Microbial ecology 🚢 oceanography 🌊 viruses 🦠 metagenomics 🧬 first-gen 🎓
https://sites.google.com/uncc.edu/microbial-ecology-lab
An expedition to study coastal ecosystems and their response to the environment, from molecules to communities
Science Events helps you organise a scientific conference or workshop. Use our platform to organise your next event! Visit our platform at https://www.sci.events