Folddisco webserver result view update:
- Added description texts for AFDB
- Integrated TaxoView taxonomy visualization & filter by @sunjaelee.bsky.social
- Inter-residue distance clustering by DBSCAN to explore motif diversity.
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
22.07.2025 18:11 — 👍 34 🔁 14 💬 0 📌 0

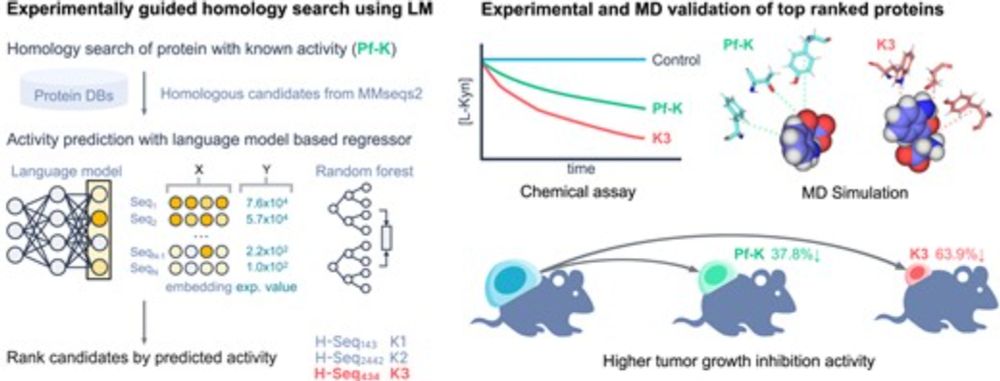
Metagenomic-scale analysis of the predicted protein structure universe
Protein structure prediction breakthroughs, notably AlphaFold2 and ESMfold, have led to an unprecedented influx of computationally derived structures. The AlphaFold Protein Structure Database now prov...
Today at 2 PM at 3DSIG #ISMBECCB2025, @nbordin.bsky.social presents our joint work on metagenomic-scale clustering and novel domain discovery in predicted structures!
📄 www.biorxiv.org/content/10.1...
Also check out poster:
B-50 lolalign Sensitive structural alignments by Lasse
B-123 BFVD by Rachel
22.07.2025 09:10 — 👍 36 🔁 8 💬 2 📌 0

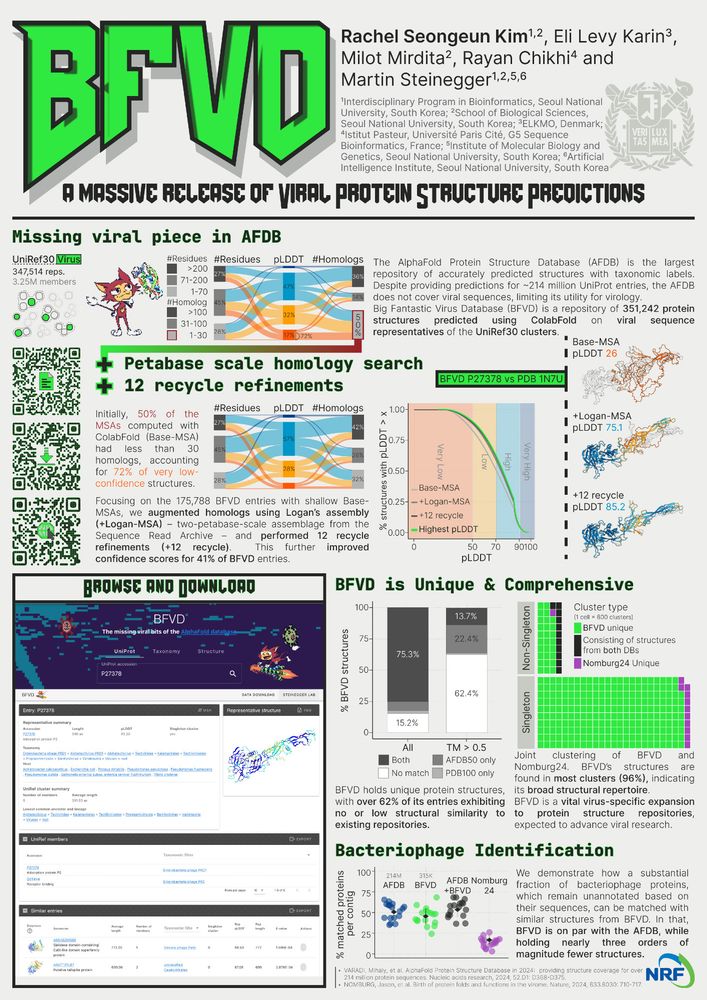
Today at 5pm, @eunbelivable.bsky.social will present her work on the Big Fantastic Viral Database (BFVD) at #ISMB2025 in BOSC. She also has a poster B-123 (tomorrow, 22nd), so please drop by to have ta chat and grab some stickers!
📄 academic.oup.com/nar/article/...
21.07.2025 09:29 — 👍 35 🔁 10 💬 1 📌 1
Thank you for the nice comments. Most figures are made with “soft” or “flat” lighting with gainsboro color for the protein :)
07.07.2025 12:50 — 👍 1 🔁 0 💬 0 📌 0
I’m excited to share our #Folddisco preprint! 🚀 We introduce a novel pairwise-geometric feature set and an optimized index structure to enable scalable structural motif search. Dive into our case studies and key results here: www.biorxiv.org/content/10.1...
07.07.2025 08:57 — 👍 7 🔁 0 💬 0 📌 0
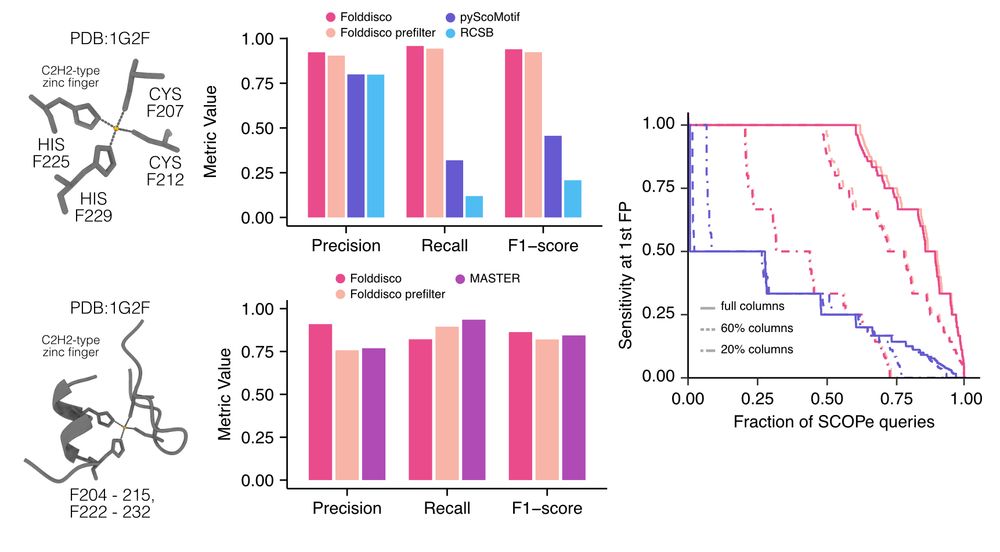
Folddisco accurately detects discontinuous motifs like zinc fingers and segment-based motifs, previously requiring separate tools. Additionally, we built a SCOPe benchmark by sampling conserved residues from families and measuring the recall up to the first false positive. 3/9
07.07.2025 08:21 — 👍 6 🔁 1 💬 1 📌 0
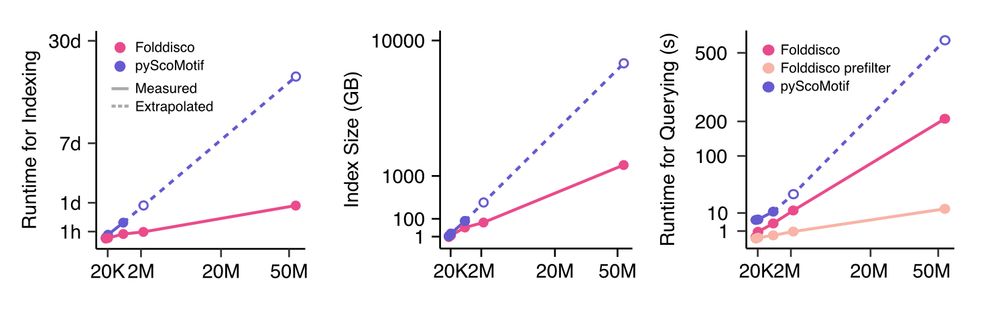
Folddisco builds indexes faster and smaller than previous tools: indexing AFDB50 (53M structures) takes only ~24h vs. ~20 days (extrapolated) for pyScoMotif. Querying a zinc-finger motif across AFDB50 takes just ~13s, up to 48x faster than pyScoMotif. 4/9
07.07.2025 08:21 — 👍 3 🔁 1 💬 1 📌 0
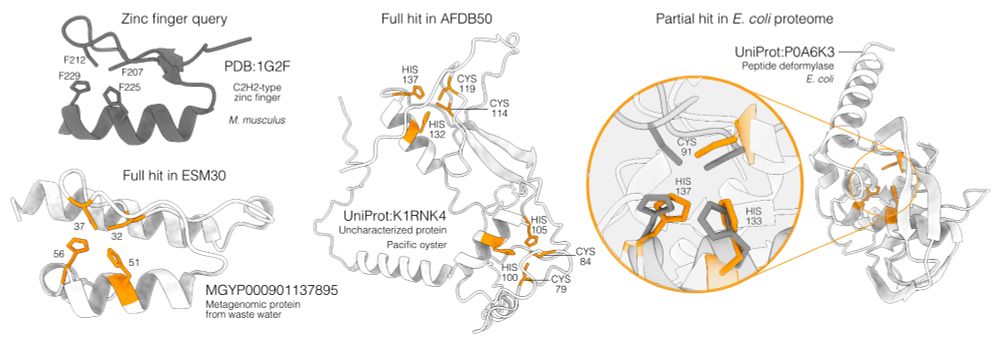
Folddisco can annotate proteins: querying a canonical zinc-finger uncovers an uncharacterized oyster protein and metagenomic proteins. It also detects partial catalytic metal sites in E. coli peptide deformylase. All of these hits would be missed by Foldseek or sequence aligners. 5/9
07.07.2025 08:21 — 👍 12 🔁 1 💬 2 📌 0
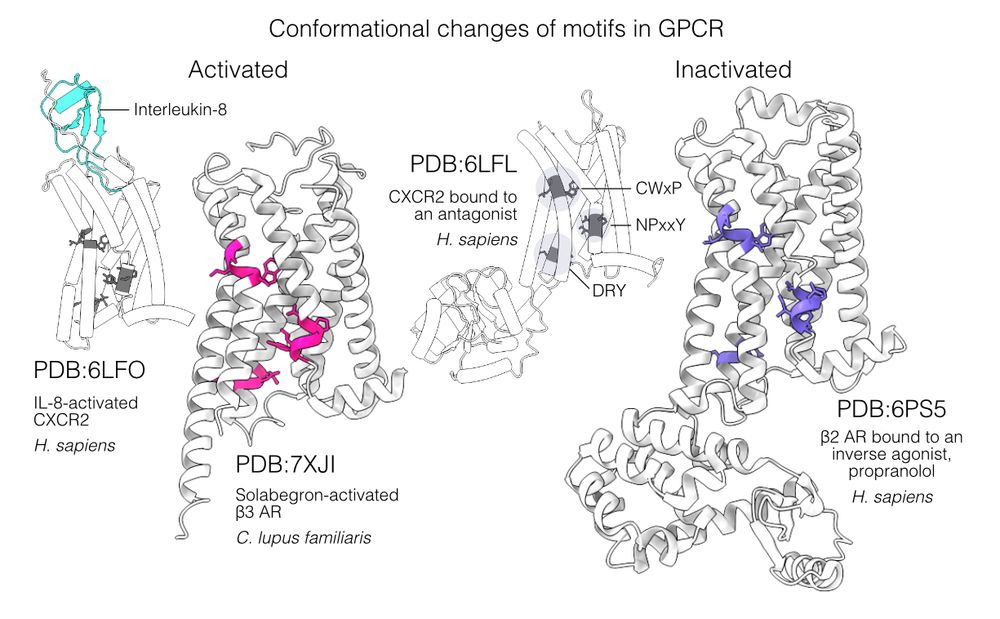
Folddisco can distinguish functional states. We searched GPCR activation motifs (CWxP, NPxxY, DRY), clearly separating active/inactive states. A search in the AFDB shows ~53% active, closely mirroring experimental PDB 54%, suggesting AlphaFold might follow its training conformation distribution. 6/9
07.07.2025 08:21 — 👍 5 🔁 1 💬 1 📌 0
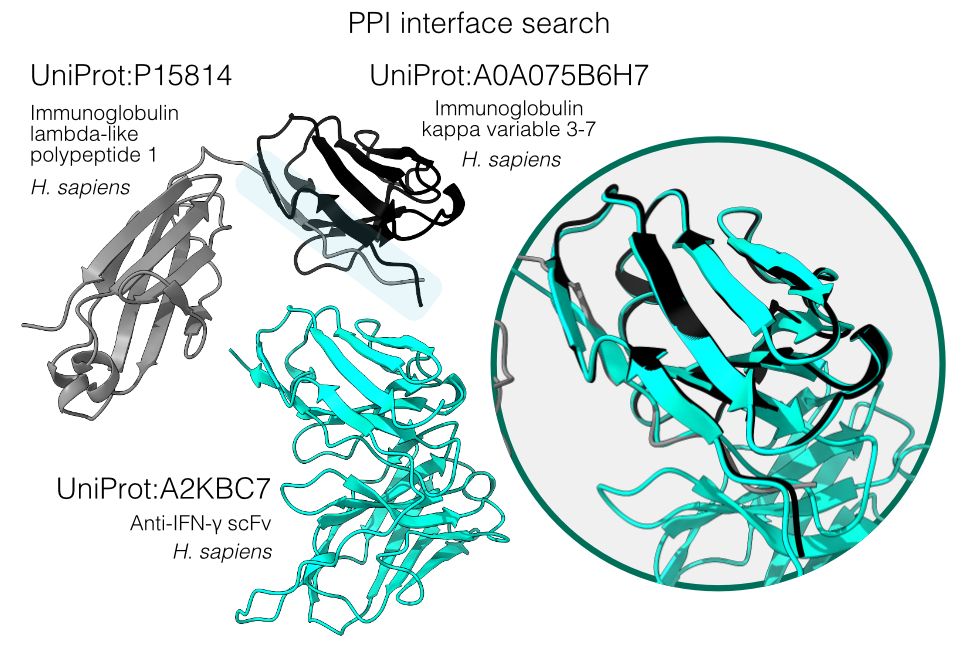
Folddisco can be applied for PPI interface searches. When querying an interface between antibody chains (gray/black), it successfully identifies matching interfaces within monomeric antibody fragments (cyan), showcasing its potential to detect novel interaction partners and interfaces. 7/9
07.07.2025 08:21 — 👍 5 🔁 1 💬 1 📌 0
We provide a user-friendly Folddisco webserver, enabling instant structural motif searches in PDB, AFDB-Proteomes, AFDB50 (available later today), and ESMatlas (ESM30). Explore it here: search.foldseek.com/folddisco 8/9
07.07.2025 08:21 — 👍 4 🔁 1 💬 1 📌 0
Structural motif search across the protein-universe with Folddisco https://www.biorxiv.org/content/10.1101/2025.07.06.663357v1
07.07.2025 03:48 — 👍 25 🔁 13 💬 0 📌 0
We've updated our AFESM website to now include biome filtering, allowing exploration of protein structures adapted to specific environments.
🌐 afesm.foldseek.com
Read more about the work in the skeetorial
🦋 bsky.app/profile/mart...
or our preprint
📄 www.biorxiv.org/content/10.1...
15.05.2025 14:03 — 👍 60 🔁 22 💬 2 📌 0
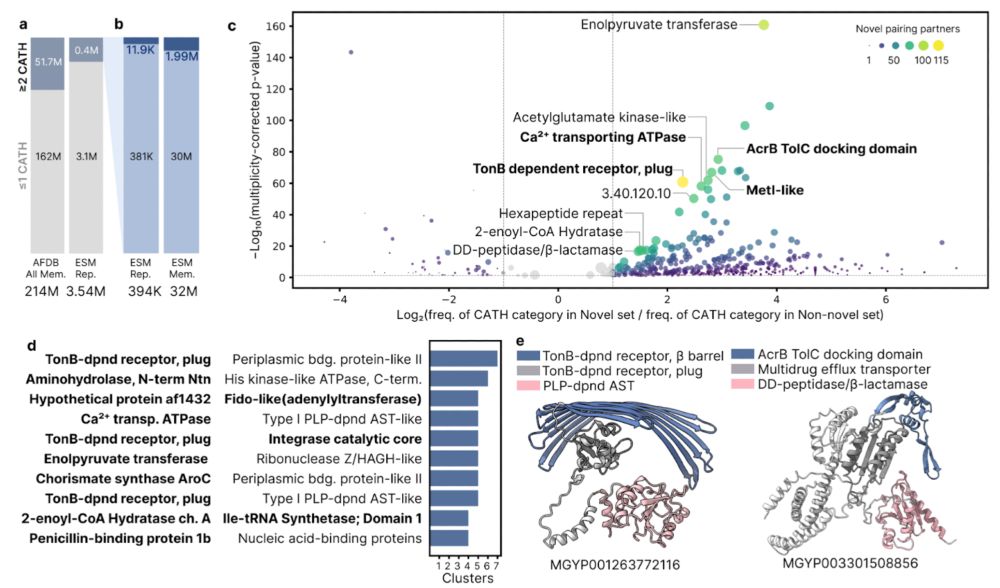
We identified 11,941 novel multi-domain combinations. We found membrane-associated domains (e.g., TonB dependent receptor, highlighting domain recombination rather than new folds as a driver of structural innovation. 5/n
27.04.2025 00:13 — 👍 9 🔁 1 💬 1 📌 0
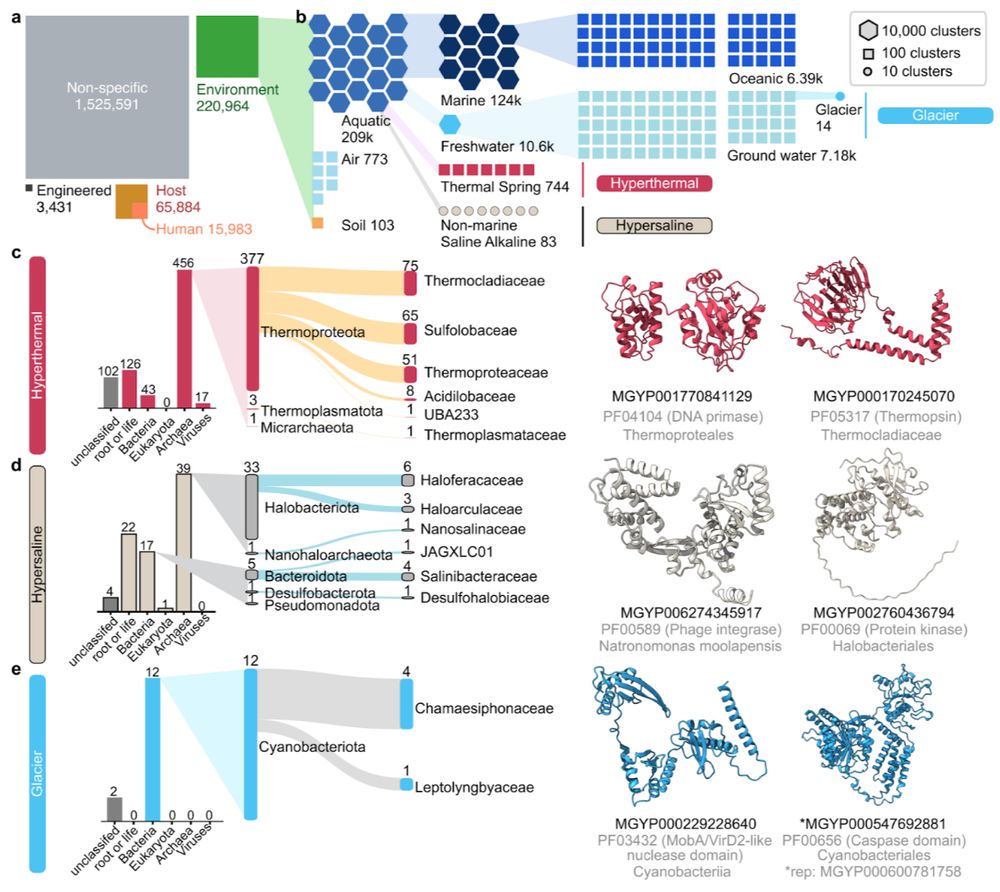
ESMatlas uses MGnify environmental labels. Leveraging this, we computed the lowest common biomes per structural cluster, revealing protein adaptations unique to specific environments, especially extreme ones like hyperthermal, hypersaline, and glaciers. 3/n
27.04.2025 00:13 — 👍 5 🔁 1 💬 1 📌 0

AFESM: a metagenomic guide through the protein structure universe! We clustered 821M structures (AFDB&ESMatlas) into 5.12M groups; revealing biome-specific groups, only 1 new fold even after AlphaFold2 re-prediction & many novel domain combos. 🧵
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
27.04.2025 00:13 — 👍 141 🔁 71 💬 4 📌 4
It's a big collaborative effort by @jingiyeo.bsky.social @yewonhan.bsky.social @nbordin.bsky.social, Andy Lau, Shaun M. Kandathil, @hbkgenomics.bsky.social, Eli Levy Karin, @milot.bsky.social David T. Jones and Christine Orengo.
Visit our #RECOMB2025 poster (719) & talk (1 pm at B145 on April 29).
27.04.2025 00:13 — 👍 10 🔁 2 💬 0 📌 0
Check out Folddisco poster at #RECOMB2025!
25.04.2025 07:54 — 👍 1 🔁 0 💬 1 📌 0
Head of Department of Algorithmics and Software,
Silesian University of Technology
bioinformatician, computer scientist
A natural product chemist using mass spec to discover chemical diversities in nature.
Working at Sookmyung Women's University, Seoul, Korea as an associate professor.
💻 We are the Structural Bioinformatics and Computational Biology track of the International Society for Computational Biology (ISCB)
PhD student at CTU Prague. Working on machine learning for molecule discovery 🤖🧪
Alzheimer research, compchem, bioinfo, neuroimaging, NBA
PhD student in structural biology with @greening.bsky.social and @knottrna.bsky.social at Monash Uni. (he/him)
Interested in hydrogenases, evolution, protein design.
💻 https://www.jameslingford.com/
Bioinformatics @UniofAdelaide @BHIresearch
- phages, microbes and more
Biologist that navigate in the oceans of diversity through space-time
Protein evolution, metagenomics, AI/ML/DL
Website https://miangoaren.github.io/
🌏 https://roman-bushuiev.github.io/
PhD student in algorithmic bioinformatics at @bonsaiseqbioinfo.bsky.social.
Interested in randomized algorithms and space-efficient data structures
https://igor.martayan.org
Computational Biologist / Bioinformatician working on novel cancer therapies. Knowledgeable in genomics, multi-omics, bioimaging, and tumour heterogeneity. Pitt-CMU CPCB alumnus. American scientist working (and cycling!) across Cambridge, UK.
Professor of Bioinformatics. Executive Editor of the Database Issue at @narjournal.bsky.social. Proud Dad. Views my own.
Trailrunner, Bioinfo Researcher at HKU D24H
linktr.ee/carloshk
Scientist @icmcsic.bsky.social #Barcelona: aquatic microbes, ecosystems, genomics, evolution, AI, bioinformatics & SciFi 🇪🇸🇦🇷 https://log-lab.barcelona
We explore the ecology and evolution of aquatic microorganisms using omics and bioinformatics. #Barcelona 🇪🇸 🇪🇺 http://log-lab.barcelona Located at @icmcsic.bsky.social
Evolutionary Virologist based at the University of Tokyo 🇯🇵
💻🦠🏳️🌈
🇵🇹 Staff Scientist @probstlab.bsky.social
(@unidue.bsky.social - 🇩🇪). Microbial genomics in the One Health context, biogeochemistry of cave microbiomes, alga-microbe symbioses.
Principal Researcher in BioML at Microsoft Research. He/him/他. 🇹🇼 yangkky.github.io
PhD | Microbiome researcher | Postdoc @Bork Group | EMBL-Heidelberg