OrthoFinder just dropped a major update
It’s faster, more accurate, and ready for thousands of genomes
Let’s break it down (1/10)
github.com/OrthoFinder/...
www.biorxiv.org/content/10.1...
16.07.2025 17:51 — 👍 126 🔁 71 💬 1 📌 1
Folddisco finds similar (dis)continuous 3D motifs in large protein structure databases. Its efficient index enables fast uncharacterized active site annotation, protein conformational state analysis and PPI interface comparison. 1/9🧶🧬
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
07.07.2025 08:21 — 👍 148 🔁 70 💬 8 📌 3

A general substitution matrix for structural phylogenetics.
Abstract. Sequence-based maximum likelihood (ML) phylogenetics is a widely used method for inferring evolutionary relationships, which has illuminated the
New paper from the lab from Sriram Garg in my group. We introduce a general substitution matrix for structural phylogenetics. I think this is a big deal, so read on below if you think deep history is important. academic.oup.com/mbe/advance-...
11.06.2025 14:01 — 👍 93 🔁 52 💬 3 📌 2
This work was done by talented @sukhwanpark.bsky.social and me, supervised by amazing @martinsteinegger.bsky.social !
Try Unicore now 👉 conda install -c bioconda unicore
Code and tutorial: 🌐 github.com/steineggerlab/unicore
Manuscript: 🌐 doi.org/10.1093/gbe/evaf109
03.06.2025 06:54 — 👍 5 🔁 0 💬 0 📌 0
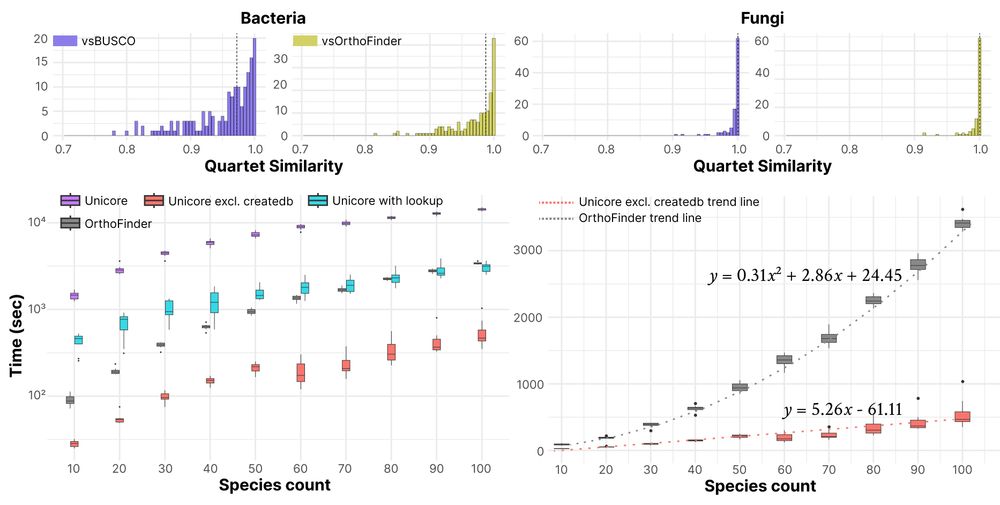
Unicore is fast, accurate, and universal. Unicore reconstructed consistent phylogeny of bacterial/fungal species, while maintaining linear time scale over the input size. Besides, Unicore works with any given taxa, presenting scalable and universal method for structure-based phylogeny. 🧵3/n
03.06.2025 06:54 — 👍 6 🔁 0 💬 1 📌 0
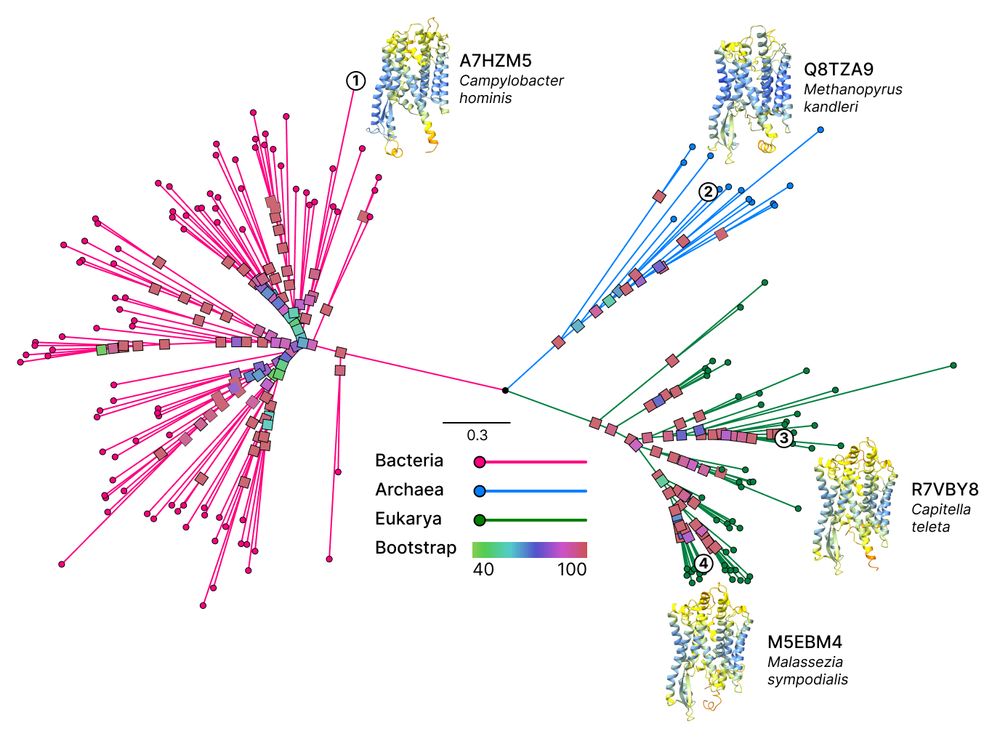
With Unicore, we identified 13 structural core genes from 166 species across the Tree of Life, where 8 of them could only be defined using structures. Projected on the Tree of Life reconstructed with Unicore, you can see the universally conserved structure of one of the structural core genes. 🧵2/n
03.06.2025 06:54 — 👍 5 🔁 0 💬 1 📌 0
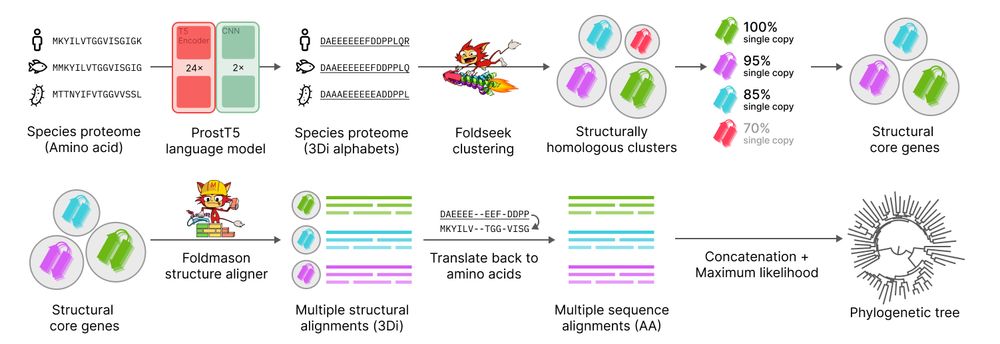
Unicore is now published on GBE 🚀
Unicore rapidly identifies structural single-copy core genes from input species proteomes for phylogenetic analysis. Powered by Foldseek and ProstT5, Unicore enables linear-scale structure-based phylogeny of any given set of taxa. 🧵1/n
📃 doi.org/10.1093/gbe/evaf109
03.06.2025 06:54 — 👍 68 🔁 31 💬 3 📌 2

AFESM: a metagenomic guide through the protein structure universe! We clustered 821M structures (AFDB&ESMatlas) into 5.12M groups; revealing biome-specific groups, only 1 new fold even after AlphaFold2 re-prediction & many novel domain combos. 🧵
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
27.04.2025 00:13 — 👍 141 🔁 71 💬 4 📌 4
Visit our posters at #RECOMB2025 for:
Structural: MSAs, Virus DB, Core Genes, Motif Discovery, Multimer Clustering & Search, pLM Foldseek, Environmental analysis
Metagenomics: Classification & Metabuli App
GPU-based & RNA search, Proteome clustering, Novel Ribozyme discovery
& get Marv stickers!
25.04.2025 07:45 — 👍 63 🔁 19 💬 2 📌 4
IQ-TREE 3: Phylogenomic Inference Software using Complex Evolutionary Models
Not really my announcement to make--I am but a lesser co-author--but IQ-TREE 3 has just been released!
(Most credit to Minh Bui and @roblanfear.bsky.social and their labs)
ecoevorxiv.org/repository/v...
10.04.2025 14:13 — 👍 179 🔁 96 💬 2 📌 6
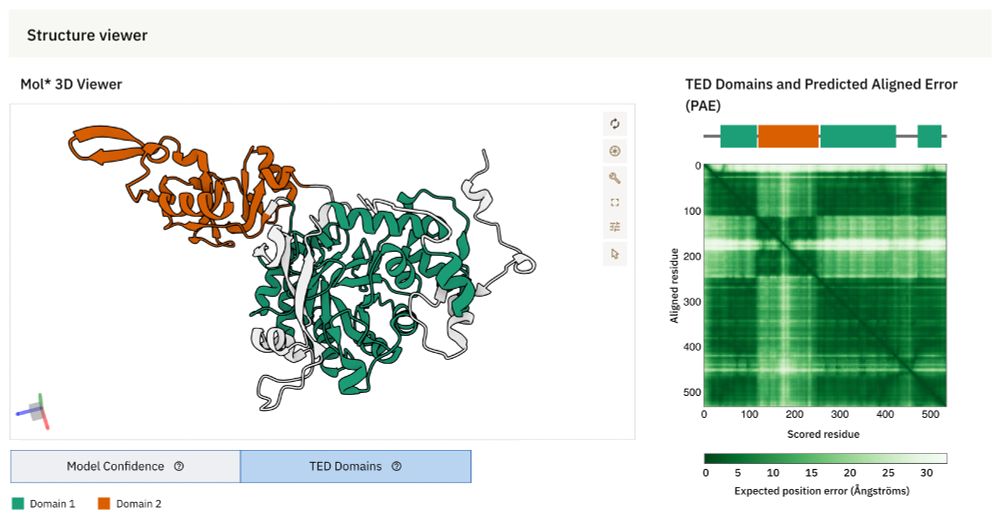
🚀 #AlphaFold Database update
AlphaFold DB now integrates The Encyclopedia of Domains (TED) – a resource designed to systematically identify & classify structural domains within AlphaFold-predicted protein structures.
www.ebi.ac.uk/about/news/u...
@pdbeurope.bsky.social
03.03.2025 16:33 — 👍 118 🔁 44 💬 1 📌 2
The PAN-GO paper is a remarkable milestone. It not only provides the most comprehensive picture of human gene function to date, but also carefully maps this knowledge across the tree of life! Congratulations @marcfeuermann.bsky.social, Pascale Gaudet & collaborators!
www.sib.swiss/news/sib-hel...
26.02.2025 22:37 — 👍 16 🔁 12 💬 0 📌 0
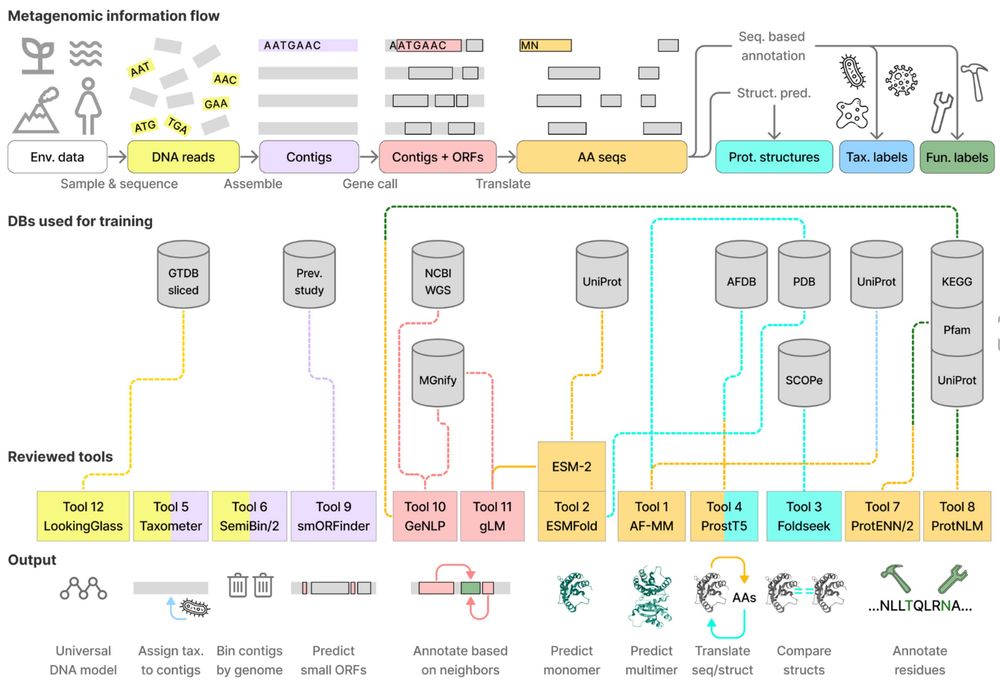
In our latest review, we explore 12 deep-learning tools for metagenomic analysis, covering their strengths, limitations, and key applications. We hope it serves as both a resource and inspiration for new ways to analyze metagenomic data. Great work by Eli Levy Karin!
📄 doi.org/10.1093/nsr/...
22.02.2025 05:47 — 👍 106 🔁 44 💬 2 📌 1
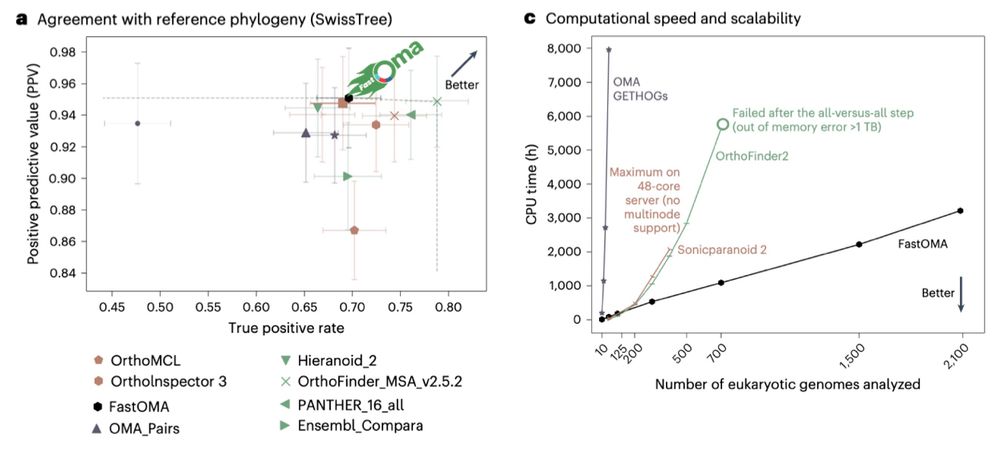
FastOMA retains OMA’s high precision accuracy and even improves upon it in terms of recall, positioning it on the Pareto frontier of orthology inference methods.
FastOMA is not only fast but also accurate. a, QfO benchmar, agreement with SwissTree reference phylogeny covering manually curated gene trees. The error bars indicate 95% confidence intervals comparing FastOMA with EnsemblCompara, Domainoid, OrthoMCL, Ortholnspector, sonicparanoid, PANTHER, OrthoFinder, Hieranoid26 and the OMA family including OMA pairs, OMA groups and OMA GETHOGs (graph-based efficient technique for HOGs).
c) A computation time comparison of FastOMA and state-of-the-art alternatives.
https://www.nature.com/articles/s41592-024-02552-8
FastOMA is out now in Nature Methods 🎉: nature.com/articles/s41592-024-02552-8 A new orthology inference algorithm that scales linearly and is highly accurate. FastOMA can process all >2000 eukaryotic UniProt ref proteomes <24 hours 🚀. Try it out github.com/DessimozLab/fastoma @dessimoz.bsky.social
03.01.2025 14:14 — 👍 35 🔁 18 💬 1 📌 0
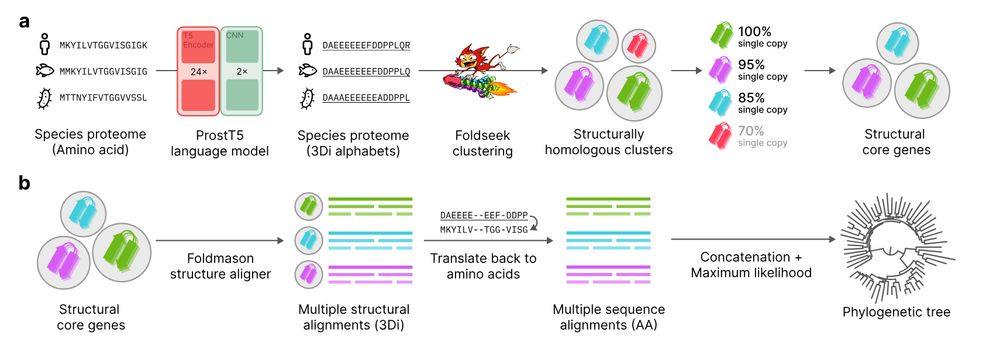
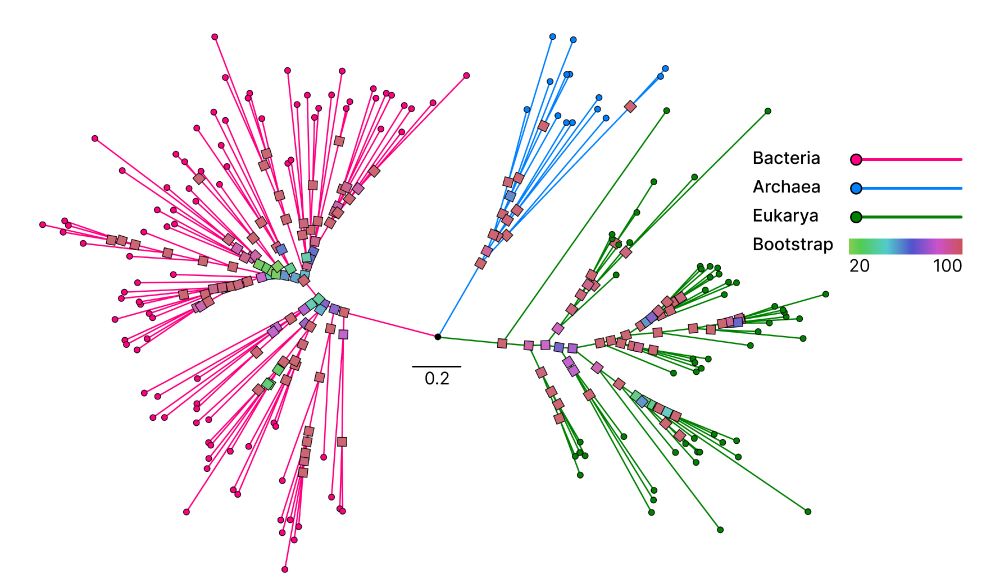
Unicore identifies single-copy protein structures across genomes using Foldseek, bypassing slow structure predictions by utilizing 3Di predictions from ProstT5, enabling rapid phylogenetic inference at the tree-of-life scale. 1/n
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...
23.12.2024 16:39 — 👍 121 🔁 57 💬 2 📌 3
Unicore enables scalable and accurate phylogenetic reconstruction with structural core genes https://www.biorxiv.org/content/10.1101/2024.12.22.629535v1
23.12.2024 03:51 — 👍 5 🔁 3 💬 0 📌 0
Scientists, academics, researchers: We’re excited to share that @altmetric.com is now tracking mentions of your research on Bluesky! 🧪
03.12.2024 14:10 — 👍 29990 🔁 5096 💬 466 📌 281
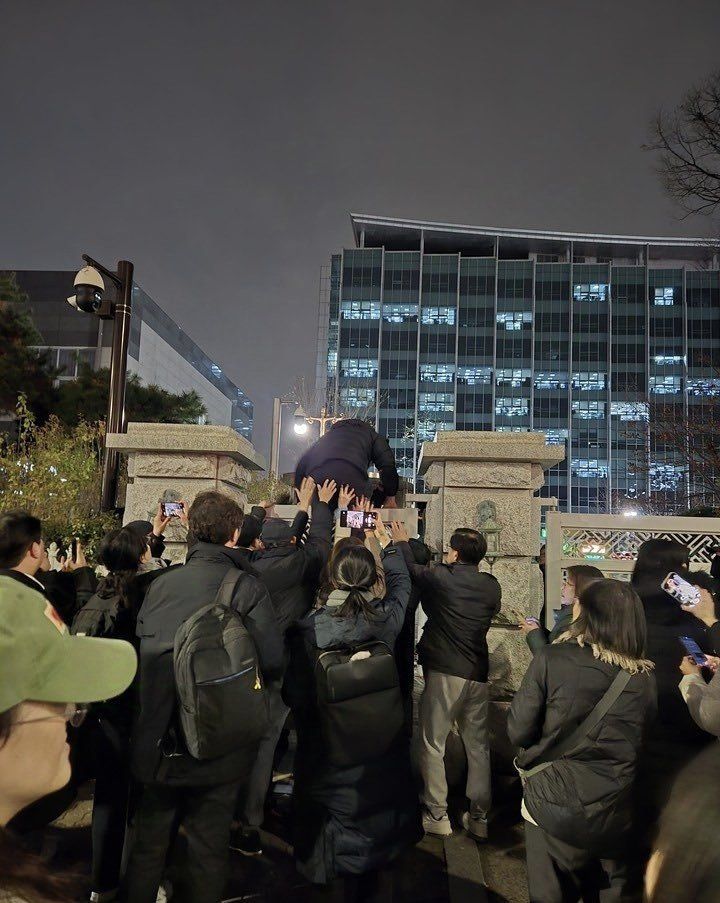
South Korean citizens helped lawmakers scale the National Assembly walls so they could bypass military barricades and vote against martial law.
03.12.2024 17:15 — 👍 13733 🔁 3178 💬 82 📌 431
Interested in bioinformatics method development for proteins, structures or metagenomic analysis? Please check out my lab’s starter pack!
🔗 go.bsky.app/VJhXcSs
28.11.2024 12:36 — 👍 56 🔁 11 💬 3 📌 0

Promotional logo for MMseqs2 16 with the MMseqs2 Rocket mascot as a smart phone like App logo
MMseqs2 Release 16 Highlights: GPU-accelerated search📄, ORF or new 6-frame translated search modes, contig taxonomy always keeps the longest ORF, bug fixes (reduced memory and higher sensitivity) and relicensed as MIT
📄 biorxiv.org/content/10.1...
💾 mmseqs.com and 🐍Bioconda 🖥️🧬🧶
27.11.2024 09:08 — 👍 116 🔁 44 💬 0 📌 1
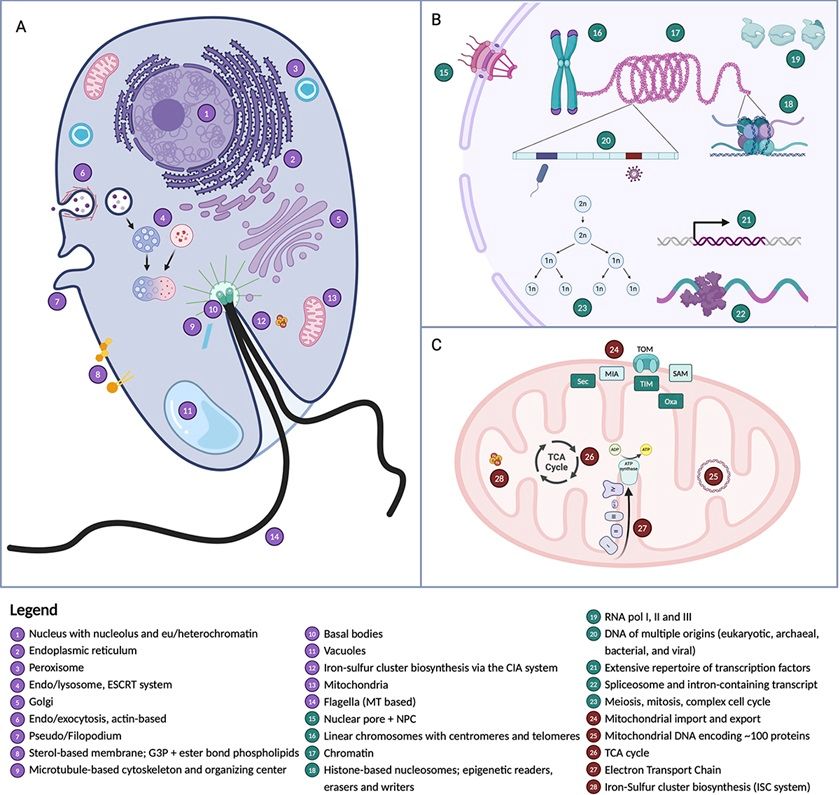
What did the Last Eukaryotic Common Ancestor (#LECA) look like? Consensus View in #PLOSBiology; massive authorship including @AncestralState, @lauraeme.bsky.social, John Archbald, @andrewjroger.bsky.social, @dackslabecb.bsky.social, Jeremy Wideman. plos.io/4g0alq4
25.11.2024 19:29 — 👍 255 🔁 101 💬 6 📌 14
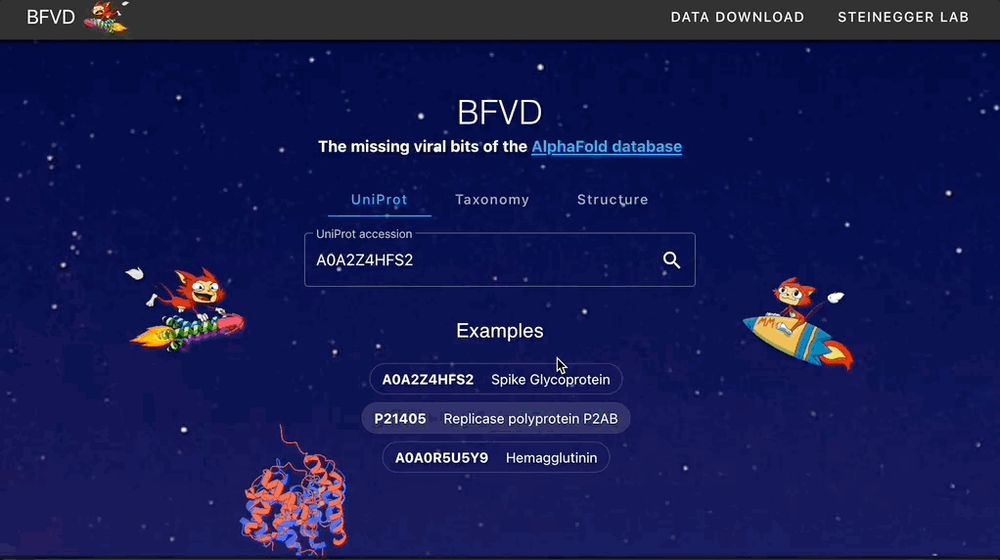
Our Big Fantastic Virus Database (BFVD) is now published NAR! It contains protein structure predictions of major viral clades, enhanced by petabase-scale homology search and it's explorable on the web.
🌐 bfvd.foldseek.com
💾 bfvd.steineggerlab.workers.dev
📄 academic.oup.com/nar/advance-...
23.11.2024 21:12 — 👍 339 🔁 127 💬 6 📌 5
👨🔬 Predoctoral Fellow at @EMBL.org as a member of the @borklab.bsky.social.
💻🧬 Using data science to analyze big bio-data.
🇪🇺 Part of the @embltrec.bsky.social expedition.
Postdoc @BorkLab @embl. PhD @UniHohenheim. Microbiome, FMT, Strain-level Metagenomics, Nutrition, ML
Data Scientist Generative AI @BayerCropScience. ML for Plant Biology. PhD @IowaStateUniversity https://www.linkedin.com/in/koushik-nagasubramanian/
Evolutionary biochemist and protein complex enthusiast at the University of Marburg.
Bloomberg Distinguished Professor of BME, CS, and Biostats at Johns Hopkins Univ., tennis player, @StevenSalzberg1 on Twitter, lab: salzberg-lab.org, Substack blog: stevensalzberg.substack.com
Biodiversity genomics; reproduction; k-mers; Evolution of species with weird genomes; frequently featuring diptera and collembola; group leader at the Tree of Life, Wellcome Sanger Institute; dysorthographic
Group leader at @ibe-barcelona.bsky.social (@csic.es - @upf.edu) & Adjunct at @miamirosenstiel.bsky.social. Microbial Ecology & Evolution 🔬🌍🧫🧬 🪸 @pr2-database.bsky.social Coordinator #corals #symbiosis #protists
Molecular Principles of RNA Phages | Junior Professor @uni-wuerzburg.de | Group Leader @helmholtz-hiri.bsky.social
Bioinformatics and marine microbial ecology. Right now focused mainly protists + metatranscriptomics! Open science. He/Him/His.
Evolutionary cell biology @EMBL
evonuclab.org
Exploring the great microbial eukaryote diversity and evolution, one protist at a time. I’m fascinated by this question: how have #plastids evolved? #ProtistsOnSky | Associate Prof at Uppsala University. My lab: https://www.burki-lab.net/
Researcher at The Sainsbury Laboratory,
Evolution | Population genetics | Plant-microbe interactions
https://yusugihara.github.io/
Bioinformatics, genomics, metagenomics, nextflow and sequencing.
💼 Junior Group Leader at CNIO @cniostopcancer.bsky.social
🔬Designing proteins and studying their structure
#cryoEM #proteindesign #ai #ml
🎓 Alumni @ UCLA, John Innes Centre, UAB
🌍 Lab website: https://www.rcglab.com
Computational Biology-Chemistry-Medicine Postdoc at University of Zurich, she/they
Metagenomicist @onecodex.bsky.social. My cat’s name is Nancy
🦠👩🏻💻Computational Virologists at the @simonlorierelab.bsky.social, the Institut Pasteur @pasteur.fr 🇫🇷, Virus Discovery & Evolution, Metagenomics 🖥️🔬🧬🦠
Website: https://xin-hou.academicwebsite.com/
🇵🇹 Staff Scientist @probstlab.bsky.social
(@unidue.bsky.social - 🇩🇪). Microbial genomics in the One Health context, biogeochemistry of cave microbiomes, alga-microbe symbioses.