Glad that we could contribute to this very cool work by @yehlincho.bsky.social and @sokrypton.org! See the nice skeetorial below by Yehlin. More to come soon with experimental validation and exciting applications in protein and peptide design.
13.10.2025 17:53 — 👍 2 🔁 0 💬 0 📌 0
We are very excited to announce that early bird registration for European RosettaCon 2025 is now open!
More information here: europeanrosettacon.org
30.06.2025 13:56 — 👍 6 🔁 2 💬 0 📌 0
Thank you!! Look forward to meeting you in Grenoble soon.
27.05.2025 01:38 — 👍 0 🔁 0 💬 0 📌 0
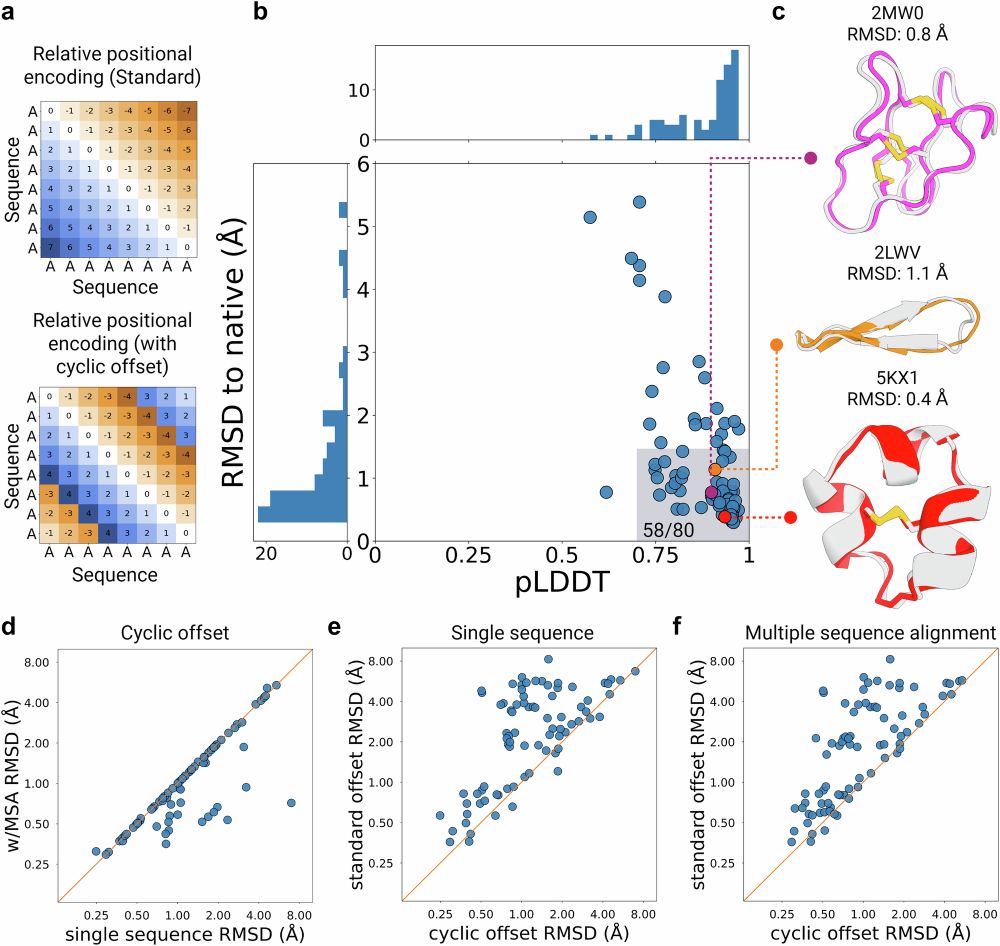
Cyclic peptide structure prediction and design using AlphaFold2
Nature Communications - AfCycDesign: Cyclic offset to the relative positional encoding in AlphaFold2 enables accurate structure prediction, sequence redesign, and de novo hallucination of cyclic...
Great to have our manuscript with @sokrypton.org 's lab describing AfCyDesign finally out in @natcomms.bsky.social . Structure prediction, sequence redesign, de novo hallucination of cyclic peptides, and some binder design examples in this version.
rdcu.be/em0vA
23.05.2025 19:07 — 👍 27 🔁 5 💬 0 📌 1
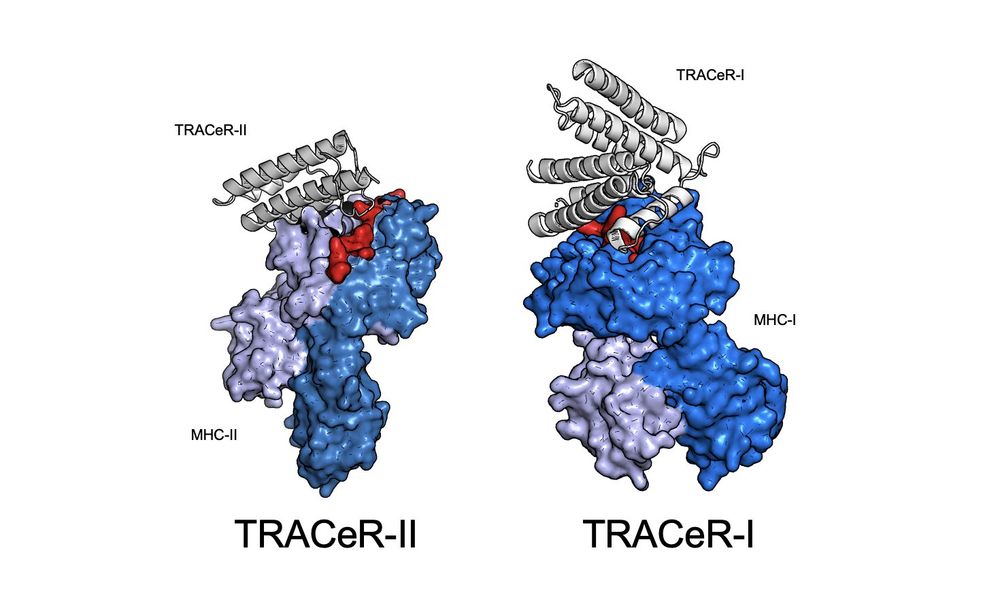
1/ In two back-to-back papers, we present our de novo TRACeR platform for targeting MHC-I and MHC-II antigens
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
17.12.2024 00:56 — 👍 92 🔁 29 💬 2 📌 9

Hans Ellegren Welcoming David Baker at Stockholm Arlanda.
Today, several #NobelPrize Laureates arrive in Stockholm, warmly welcomed by Hans Ellegren. Here we see David Baker stepping off the plane at Arlanda.
This week is packed with inspiration, press conferences and lectures, so stay tuned! 🌟
@uofwa.bsky.social @hhmi.bsky.social
#Science #AcademicSky
05.12.2024 12:48 — 👍 74 🔁 15 💬 0 📌 1
Thanks for the shoutout! I agree - We have not tried it, but
don't expect it to work well for disordered targets with low-throughput testing. Not yet, at least! 😀
21.11.2024 17:57 — 👍 0 🔁 0 💬 0 📌 0
Hats off to the people compiling all those starter packs—it has made the move to this site so much easier!
21.11.2024 17:52 — 👍 5 🔁 0 💬 0 📌 0
Ha ha Probably not! :)
21.11.2024 06:28 — 👍 0 🔁 0 💬 0 📌 0
I may have figured out how to add a GIF of diffusion trajectory. Lets see! 😀
i.giphy.com/media/v1.Y2l...
21.11.2024 06:21 — 👍 0 🔁 0 💬 1 📌 0
It is a really fun time to be designing peptides/proteins. Please reach out if you have targets you would like to design macrocycle binders.
21.11.2024 06:21 — 👍 0 🔁 0 💬 1 📌 0
None of this would have been possible without all the great collaborators at @uwproteindesign.bsky.social and beyond (still trying to find everyone here!). There is much more to come as we continue to fine-tune and expand RFpeptides.
21.11.2024 06:21 — 👍 0 🔁 0 💬 1 📌 0
Perhaps the most fun part for us was the RbtA, where we did not have the target structure available when we designed against it. So we predicted the structure using AF2/RF2 and then designed against the predicted structures. Tested < 15 designs and got a Kd <10 nM binder!
21.11.2024 06:21 — 👍 1 🔁 0 💬 1 📌 0
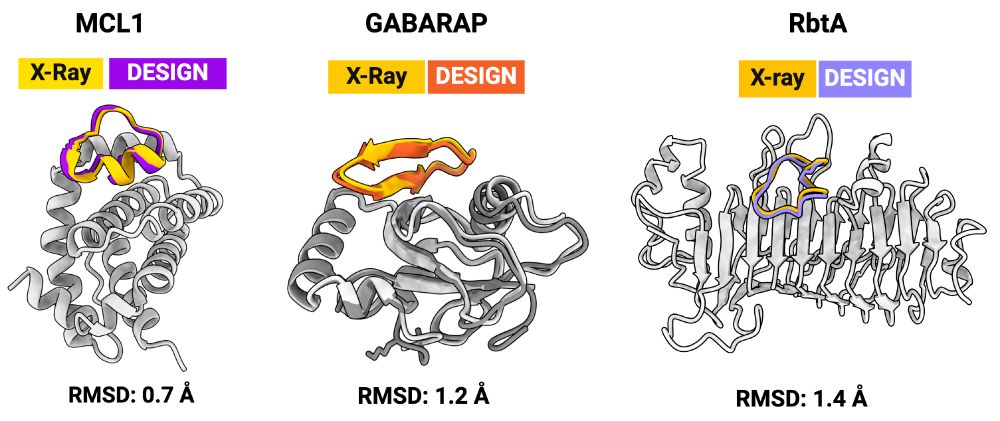
X-ray structures for the macrocycle bound complexes also match very closely with the design models (CA RMSD < 1.5 angstroms). The designs are diverse: helix-containing (MCL-1/Mdm2), beta-strands (GABARAP), and loopy (RbtA).
21.11.2024 06:21 — 👍 0 🔁 0 💬 1 📌 0
We used RFpeptides to design binders against four different targets: Mdm2, MCL-1, GABARAP, and RbtA. For each of the targets, we experimemtally tested <20 designs. We got 1-10 micromolar Kd binders against Mdm2 and MCL-1, and 1-10 nM binders against GABARAP and RbtA.
21.11.2024 06:21 — 👍 0 🔁 0 💬 1 📌 0
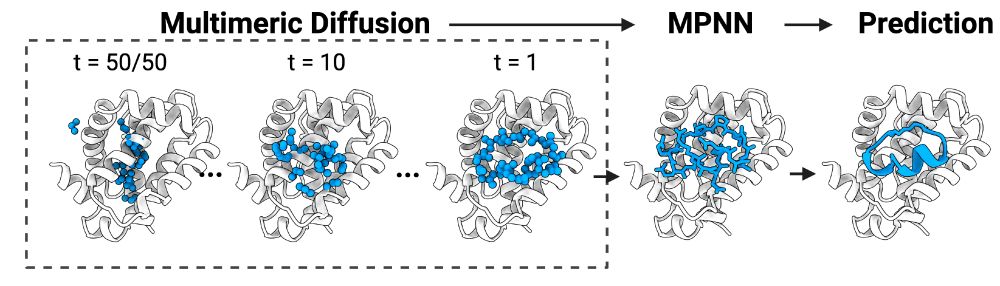
Here, we modified RFdiffusion positional encodings to design cyclic peptide backbones against selected targets, followed by sequence design using ProteinMPNN. Final designs were selected based of confidence metrics from AF2/RF2 re-prediction and Rosetta-based interface quality metrics.
21.11.2024 06:21 — 👍 0 🔁 0 💬 1 📌 0
So the design pipeline has to be good at designing and also selecting the best 10-20 binders. And it should also work for diverse targets. RFpeptides seems to be able to address a lot of those early issues and meet the requirements.
21.11.2024 06:21 — 👍 0 🔁 0 💬 1 📌 0
Why are we excited about it? Well, we spent a lot of effort over the years to accurately design high-affinity binders with our physics-based methods without much success. Since we rely on chemical synthesis of macrocycles, we were limited to making and testing only 10-20 designs/target in our lab.
21.11.2024 06:21 — 👍 0 🔁 0 💬 1 📌 0
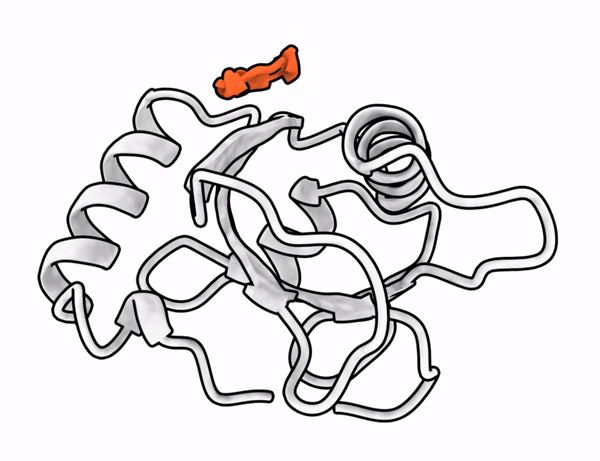
Here goes the skeetorial for the latest preprint from our lab describing RFpeptides, a pipeline for design of target-binding macrocycles using diffusion models. Big shoutout to Stephen Rettie, David Juergens, Victor Adebomi for leading the project (1/n)
Preprint link: www.biorxiv.org/content/10.1...
21.11.2024 06:21 — 👍 15 🔁 8 💬 2 📌 1
Here is a GIF in the meantime:
19.11.2024 19:08 — 👍 2 🔁 0 💬 0 📌 0
PhD researcher @ University of Copenhagen & Gubra
Machine Learning for design and optimization of peptide therapeutics
CEO at Vilya @vilyatx.bsky.social where we use AI to design novel macrocycles. Founder, board member, and ex-CEO at Olema where we design and develop new drugs for breast cancer.
Your complete source for all the life science news and events in the Seattle area. Brought to you by the Science in the City program @stemcell.com.
Staff scientist @biozentrum.unibas.ch using algorithms to look at tiny stuff inside cells, or #cryoET if you prefer. Here I talk mostly about my research. Views my own. 🇧🇷 in 🇨🇭
Researcher in computational biology / bioinformatics at @i2bcparissaclay.bsky.social 🇫🇷
Protein-protein, protein-RNA & protein-DNA interactions, structure & evolution
ML & DL
Biology of genome maintenance
Women/diversity in science
Personal account
Group Leader at EMBL Grenoble. mRNA, tRNA, RNA modifications, structural biology, x-ray, cryo-EM, trypanosomes and other science fun.
Professor of Theoretical Chemistry @sorbonne-universite.fr & Head @lct-umr7616.bsky.social| Co-Founder & CSO @qubit-pharma.bsky.social| FRSC (My Views) #compchem #HPC #quantumcomputing #machinelearning |
https://piquemalresearch.com | https://tinker-hp.org
Nature Communications is an open access journal publishing high-quality research in all areas of the biological, physical, chemical, clinical, social, and Earth sciences.
www.nature.com/ncomms/
We study antiviral immunity and viral disease pathogenesis. We are developing mucosal vaccine strategies to prevent infection and transmission. #COVID19 #longCOVID #vaccines
PhD student@MIT
https://sites.google.com/view/yehlincho/home
News from the King Lab kinglab.ipd.uw.edu.
Part of @uwmedicine.bsky.social and @uwproteindesign.bsky.social.
Some assembly required.
Part of the UW School of Pharmacy, researching drug metabolism, bio-analytical chemistry and the biophysical characterization of viral protein assembly.
CEO of Bluesky, steward of AT Protocol.
dec/acc 🌱 🪴 🌳
Econ professor at Michigan ● Senior fellow, Brookings and PIIE ● Intro econ textbook author ● Think Like An Economist podcast ● An economist willing to admit that the glass really is half full.
Congresswoman, lifelong organizer, mom. Proudly serving WA-7. Chair Emerita of the Congressional Progressive Caucus. Member of Judiciary Committee, Foreign Affairs Committee, Budget Committee. She/her.
Assistant Professor @ucf.bsky.social
https://medvedev-lab.cs.ucf.edu/
Alumni @utswtxid.bsky.social
Computational Biology | Bionformatics | Cancer Biology
Freelance writer, editor, columnist & consultant. Formerly at Scientific American, Washington Post, National Geographic, Slate, Smithsonian & Science. Past president of National Association of Science Writers. Birder.
Husband, dad, veteran, writer, and proud Midwesterner. 19th US Secretary of Transportation and former Mayor of South Bend.
Host of the Fast politics podcast, contributing Writer at The New York Times opinion and MSNBC. Subscribe to fast politics YouTube channel







