🎉 Congrats to Dr. Laura Luebbert ( @lauraluebbert.com ) for receiving the 2025 FutureHouse Fellowship! She’ll develop AI tools to detect viral sequences in genomic data & analyze virome data from 4,000+ people. Creator of gget & champion of open science—go Laura! 🧬🙌
14.05.2025 14:43 — 👍 6 🔁 3 💬 1 📌 0
Very excited to finally see this out! 🦠🧬
Huge thanks to all of my amazing co-authors! @lpachter.bsky.social @delaneyksull.bsky.social
Original paper thread: x.com/neuroluebber...
Free access link to the paper: rdcu.be/eiIWC
22.04.2025 14:15 — 👍 28 🔁 11 💬 0 📌 0
Introduction - gget
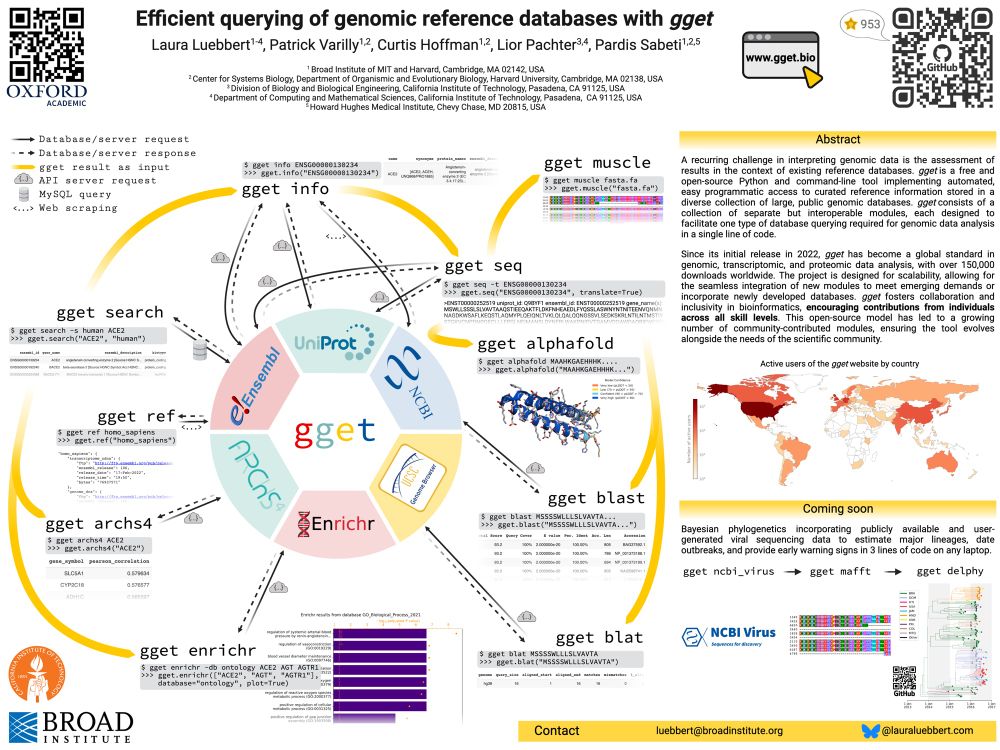
gget enables efficient querying of genomic reference databases
Does it come with an API? We could use this under the hood of gget blast as well
pachterlab.github.io/gget/
12.03.2025 14:03 — 👍 3 🔁 0 💬 0 📌 0
gget hit >1,000 stars on GitHub!!! 🤯
Very grateful to all gget users and contributors for continuously helping us make bioinformatics databases and tools more accessible! ✨
github.com/pachterlab/g...
12.03.2025 13:57 — 👍 8 🔁 1 💬 1 📌 0
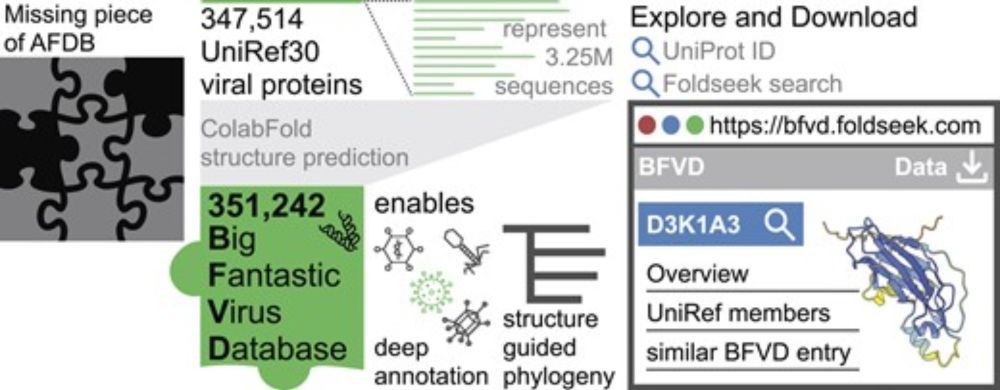
BFVD—a large repository of predicted viral protein structures
Abstract. The AlphaFold Protein Structure Database (AFDB) is the largest repository of accurately predicted structures with taxonomic labels. Despite provi
Our Big Fantastic Virus Database (BFVD) contains protein structure predictions of major viral clades, enhanced by petabase-scale homology search and it's explorable on the web. Led by @eunbelivable.bsky.social
🌐 bfvd.foldseek.com
📄 academic.oup.com/nar/article/...
08.03.2025 15:28 — 👍 5 🔁 1 💬 1 📌 0
Fantastic slides describing gget!
Thank you for sharing #HoffmanLabTechTalk @michaelhoffman.bsky.social and Luomeng Tan.
github.com/pachterlab/g...
07.03.2025 16:23 — 👍 3 🔁 2 💬 0 📌 0

The Nature briefing features my work on nests. We see one picture of the Rokin nest with a lot of plastic sticking out - prior to collecting it. And the stratigraphy of plastic with all the datable pieces of the nest laid out as a timeline.
Feels kind of illegal to be featured by "Nature" with a bird's nest this artificial. But here we are. 😅🪹🪶
@nature.com 🌿🧪
05.03.2025 20:19 — 👍 142 🔁 17 💬 10 📌 2
Still one of the coolest things we've ever done. Two degreeless heathens in a home lab published a microbial genome so clean and complete it became THE reference for that organism.
www.ncbi.nlm.nih.gov/datasets/tax...
03.03.2025 15:17 — 👍 86 🔁 4 💬 2 📌 0
Thank you!
02.03.2025 22:56 — 👍 0 🔁 0 💬 0 📌 0
I have no problem accessing NCBI from my laptop at home in Cambridge, but the server errors out as soon as I connect to any sort of VPN (including Boston VPNs) or try to access it from GitHub Actions. Anyone know what’s going on?
02.03.2025 17:36 — 👍 0 🔁 0 💬 1 📌 0

JXTX + CSHL 2025 Biology of Genomes Scholarship
JXTX + CSHL 2025 Biology of Genomes Scholarship
Tomorrow is the deadline for applications for JXTX Scholarships for the CSHL Biology of Genomes! The application is lightweight - just a couple brief statements about your work! @cshlmeetings.bsky.social @jxtxfoundation.bsky.social jxtxfoundation.org/news/2025-2-...
11.02.2025 21:49 — 👍 11 🔁 13 💬 0 📌 1

It’s been a tough few weeks. My 10yo daughter was diagnosed with a very rare, aggressive cancer called interdigitating dendritic cell sarcoma (IDCS). I’m reaching out to identify clinicians/patients who have encountered pediatric IDCS or other (non-LCH) dendritic or histiocytic sarcomas cases.
08.02.2025 21:21 — 👍 1017 🔁 863 💬 82 📌 32
Wondering which diseases or drugs are associate with your gene of interest (including ongoing clinical trials)?
You can now answer this question directly from your command-line or Python/R environment:
pachterlab.github.io/gget/en/open...
Huge shoutout to Joseph Rich and Sam Wagenaar!
30.01.2025 15:23 — 👍 7 🔁 1 💬 0 📌 0

Case study: gget’s new Open Target module
The new gget opentargets module allows users to communicate directly with the Open Targets database from a Python or command line environment. Amongst other tasks, gget opentargets can quickly find di...
With the gget opentargets module, you can now interact directly with the Open Targets database from a Python or command line environment
Developed by @lauraluebbert.com, gget is a free, open-source tool enabling efficient querying of large genomic databases 🖥️🧬
blog.opentargets.org/case-study-g...
30.01.2025 11:56 — 👍 10 🔁 5 💬 0 📌 1
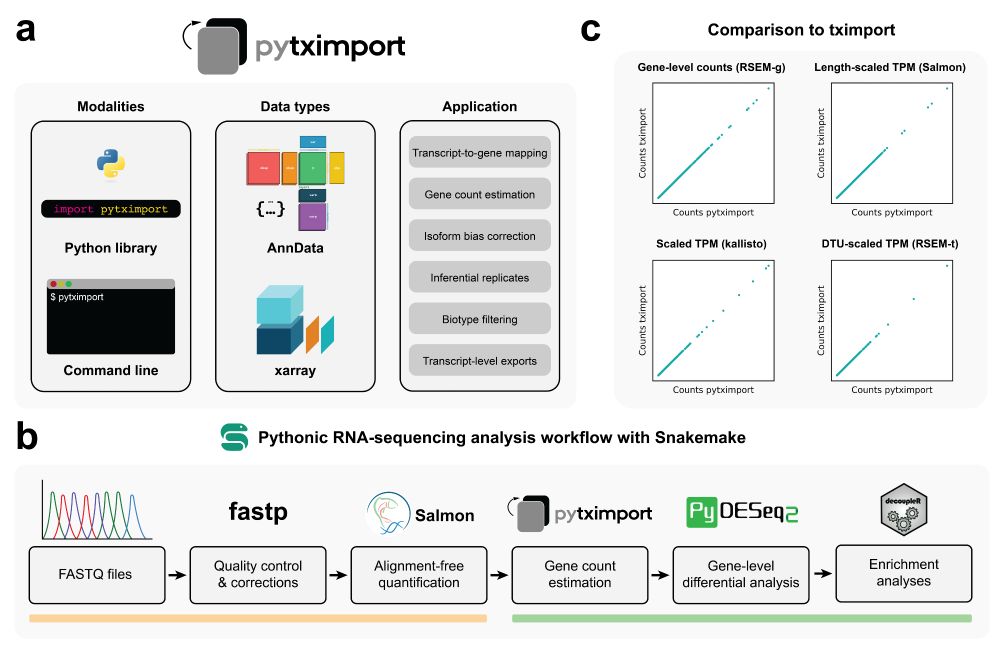
Figure 1 from the pytximport manuscript. Overview of the pytximport package and its associated RNA sequencingworkflow. a) pytximport package. pytximport is available for use as a Python library orfrom the command line. It can be configured to either output AnnData objects forintegration with other scverse ecosystem software or xarray datasets. Common applicationsfor pytximport include gene count estimation from transcript quantification files, isoform-usage bias correction, filtering of transcript-level data and creation of transcript-to-genemappings. b) Pythonic RNA sequencing analysis workflow. We propose a reproducibleRNA-seq analysis workflow based on command-line software available through Bioconda(yellow line: Snakemake, fastp, Salmon) and scverse ecosystem Python packages (greenline: pytximport, PyDESeq2, decoupleR). c) Comparison with tximport. Counts frompytximport match counts from tximport exactly across different quantification modes andinput files from different transcript quantification tools. RSEM-g: RSEM gene-level input.RSEM-t: RSEM transcript-level input.
Coding in Python and looking to perform bulk RNA sequencing analysis? With pytximport recently published in Bioinformatics and version 0.11.0 out today with many improvements, it’s time for my first Bluetorial!
A thread 🧵 (1/12)
29.11.2024 17:24 — 👍 35 🔁 14 💬 2 📌 1
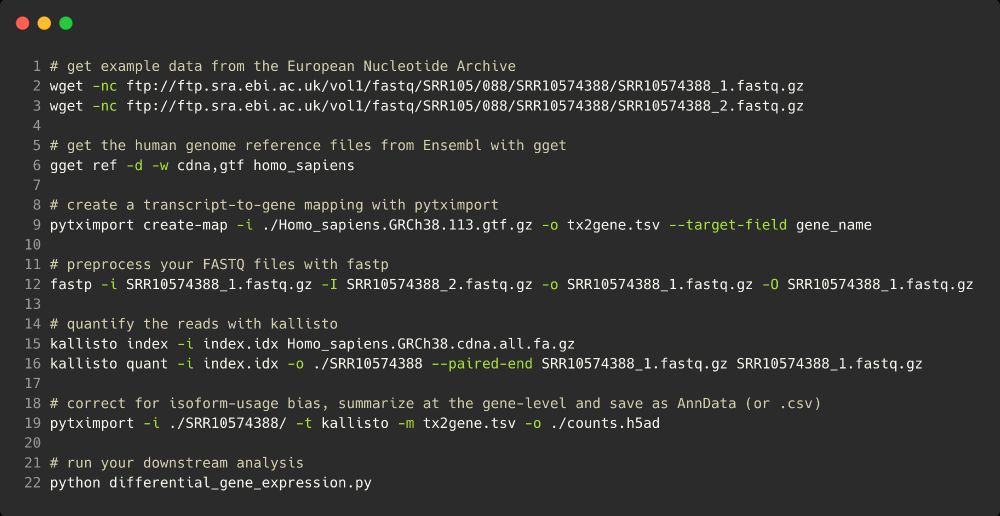
Code for RNAseq analysis:
# get example data from the European Nucleotide Archive
wget -nc ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR105/088/SRR10574388/SRR10574388_1.fastq.gz
wget -nc ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR105/088/SRR10574388/SRR10574388_2.fastq.gz
# get the human genome reference files from Ensembl with gget
gget ref -d -w cdna,gtf homo_sapiens
# create a transcript-to-gene mapping with pytximport
pytximport create-map -i ./Homo_sapiens.GRCh38.113.gtf.gz -o tx2gene.csv --target-field gene_name
# preprocess your FASTQ files with fastp
fastp -i SRR10574388_1.fastq.gz -I SRR10574388_2.fastq.gz -o SRR10574388_1.fastq.gz -O SRR10574388_1.fastq.gz
# quantify the reads with kallisto
kallisto index -i index.idx Homo_sapiens.GRCh38.cdna.all.fa.gz
kallisto quant -i index.idx -o ./SRR10574388 --paired-end SRR10574388_1.fastq.gz SRR10574388_1.fastq.gz
# correct for isoform-usage bias, summarize at the gene-level and save as AnnData (or .csv)
pytximport -i ./SRR10574388/ -t kallisto -m tx2gene.csv -o ./counts.h5ad
# run your downstream analysis
python differential_gene_expression.py
With pytximport and tools like fastp by Shifu Chen, kallisto by the @lpachter.bsky.social lab and gget by @lauraluebbert.com et al, running a bulk RNA-sequencing analysis is now a 10 line bash script. (7/12)
29.11.2024 17:24 — 👍 2 🔁 2 💬 1 📌 0

This week was my last retreat as a postdoc at the @broadinstitute.org 🥹 I was incredibly honored to receive the Eric S. Lander award and to talk about the dark proteome of viruses. Excited to begin my next chapter at @harvardmed.bsky.social next month! (thx @lauraluebbert.com for the lovely 📸)
19.12.2024 05:06 — 👍 18 🔁 4 💬 2 📌 0

Here is a thread showcasing my GitHub repositories: 1) Friends Don't Let Friends Make Bad Graphs. An opinionated essay on good and bad graphs.
My popular one by a long shot with 6.4k stars and 248 forks. github.com/cxli233/Frie...
12.11.2024 02:40 — 👍 183 🔁 58 💬 10 📌 2
Here's a new starter pack for researchers at the Broad Institute of MIT and Harvard!
22.11.2024 01:26 — 👍 9 🔁 3 💬 5 📌 0
Could you please add me? :)
22.11.2024 01:52 — 👍 0 🔁 0 💬 1 📌 0
I've posted the notes/slides for my computational biology class at github.com/pachterlab/B... Topics were chosen based on appearing in >=3 bio areas, although for focus examples are all drawn from #scRNAseq. Homeworks include both theory and exploration of data (via GoogleColab).
22.09.2023 13:55 — 👍 87 🔁 23 💬 2 📌 2
The Journal of Scientific Integrity
by Laura Luebbert and Lior Pachter Background (by LL) Four years ago, during the first year of my PhD at Caltech, I participated in a journal club organized by the lab I was rotating in. I was assi…
A friend mentioned honey bee waggle dance to me recently and I had to tell them the bad news about that literature: numerous key papers appear to be fraud. So here is me sharing the bad news with you too. Extensive blog post by the hero sleuth, Laura Luebbert, and @lpachter.bsky.social:
19.11.2024 08:28 — 👍 193 🔁 49 💬 7 📌 2
Great idea @lpachter.bsky.social! In the meantime, I built a Chrome extension that puts a share to Bluesky button next to the X button on any @biorxivpreprint.bsky.social preprint. Installation instructions in the repo github.com/stephenturne.... I'll get this on the Chrome web store soon. 🧬🖥️🧪
20.11.2024 14:07 — 👍 56 🔁 13 💬 1 📌 3
I'm delighted to share that I will be joining the Department of Microbiology at Harvard Medical School in January 2025. The Laboratory of Systems Virology will study the dark proteome of viral genomes to understand how viruses work their magic🪄 Thrilled to be back in beautiful Boston 🌟
20.11.2024 14:14 — 👍 186 🔁 11 💬 4 📌 2
Hola! We are the CRG, a research institute exploring the frontiers of biology in Barcelona. Som un centre CERCA. www.crg.eu
Relaying microbiology news, articles and comments relevant to aspects on bacteria, fungi, viruses and other microbes / microorganisms. Microbes are 💪
Postdoc at Helsinki University, focused on viruses and predatory bacteria in soil, permafrost, sea ice, and high salinity environments 🧫🧬
Group leader in the Virology Department at Institut Pasteur
🦠👩🏻💻Computational Virologists at the @simonlorierelab.bsky.social, the Institut Pasteur @pasteur.fr 🇫🇷, Virus Discovery & Evolution, Metagenomics 🖥️🔬🧬🦠
Website: https://xin-hou.academicwebsite.com/
Postdoctoral Scholar MIT and the Broad Institute of MIT and Harvard | Previous Caltech PhD |
We are the Melé Lab, the Transcriptomics and Functional Genomics Lab (TFGL) at the Barcelona Supercomputing Center (BSC).
Account by lab members
The official Bluesky account of the Massachusetts Bay Transportation Authority (MBTA)
mbta.com
Boston 25 News provides complete New England news coverage that matters to you.
BPS mom, daughter of immigrants, and MBTA commuter working to make Boston a home for everyone.
linktr.ee/mayorwu
Follow the @boston.gov Bluesky Starterpack: https://bsky.app/starter-pack/boston.gov/3lcvbzv3ttb25
CEO of Bluesky, steward of AT Protocol.
Let’s build a federated republic, starting with this server. 🌱 🪴 🌳
Sci journo, author of Pests: How Humans Create Animal Villains. Highly caffeinated. All bad takes mine. She/her
Computational biologist & blogger
Wir sind die Fachgesellschaft für Ärzt/Innen und Wissenschaftler/Innen aus dem Bereich der Virologie in und aus Deutschland, Österreich und der Schweiz
https://g-f-v.org/
CS PhD student at @jhu.edu @jhucompsci.bsky.social
Teaching machines to learn biology 🧬💻
https://khchao.com/
NYT columnist. Signal: carlzimmer.31
Newsletter: https://buttondown.com/carlzimmer/
Web: http://carlzimmer.com
[This account includes a tweet archive]
scientist at UC Berkeley inventing advanced genomic technologies
lover of molecules, user of computers
https://scholar.google.com/citations?user=63ZRebIAAAAJ&hl=en
Because Science is for everyone! 🧪🌎
Learn more ⬇️
http://linktr.ee/standupforscience
👯sister org: @sciforgood.bsky.social