
Join us January 22 for Proteoform Thursday: Pei Su of UC Riverside presents "From Tissues to Single Cells: Direct Proteoform Profiling Using Orbitrap-Based Single Molecule Mass Spectrometry" #proteomics #proteoform #massspec us06web.zoom.us/meeting/regi...
19.12.2025 16:35 — 👍 2 🔁 2 💬 0 📌 0
Wonderful native individual ion mass spectrometry work!
10.12.2025 21:31 — 👍 2 🔁 0 💬 0 📌 0
I want to thank @us-hupo.org for the Travel Award that made this trip possible; and #Affinisep for the generous and supportive seminar invitation.
21.11.2025 20:13 — 👍 1 🔁 0 💬 0 📌 0


It was a great pleasure attending my first #HUPO world congress in Toronto last week and sharing our catenin phospho-code story! It was inspiring meeting with peers and pioneers in the field, which reminded me how special the proteomics community is!
21.11.2025 20:13 — 👍 2 🔁 1 💬 1 📌 0
TECHNICAL INNOVATION: We also demonstrated that I2MS for fragment ion analysis (I2MS2) increases catenin top-down sequence coverages from <1% to ~30%. We expect future instrumentation advances will improve proteoform-level analysis of these LARGE structural proteins/complexes. (7/7)
31.10.2025 20:00 — 👍 2 🔁 0 💬 0 📌 0
BIOLOGY TAKEAWAY: We proposed a catenin phospho-code—where combinatorial phosphorylation of these large proteoforms works with mechanical signals to drive cadherin–catenin organization and #adherensjunction function. (6/7)
31.10.2025 20:00 — 👍 2 🔁 0 💬 1 📌 0

Using bottom-up proteomics/antibodies, we confirmed that some phosphorylations appear constitutive, while others are more sensitive to myosin inhibition, raising the possibility of a mechano-sensitive “phospho-code” for cell-cell adhesion. (5/7)
31.10.2025 20:00 — 👍 1 🔁 0 💬 1 📌 0

Remarkably, catenin phosphorylation is sensitive to drugs that modulate #actomyosin contractility. We observed HYPER-phosphorylation on catenins under conditions that promote cell rounding and HYPO-phosphorylation when myosin activity was reduced. (4/7)
31.10.2025 20:00 — 👍 1 🔁 0 💬 1 📌 0
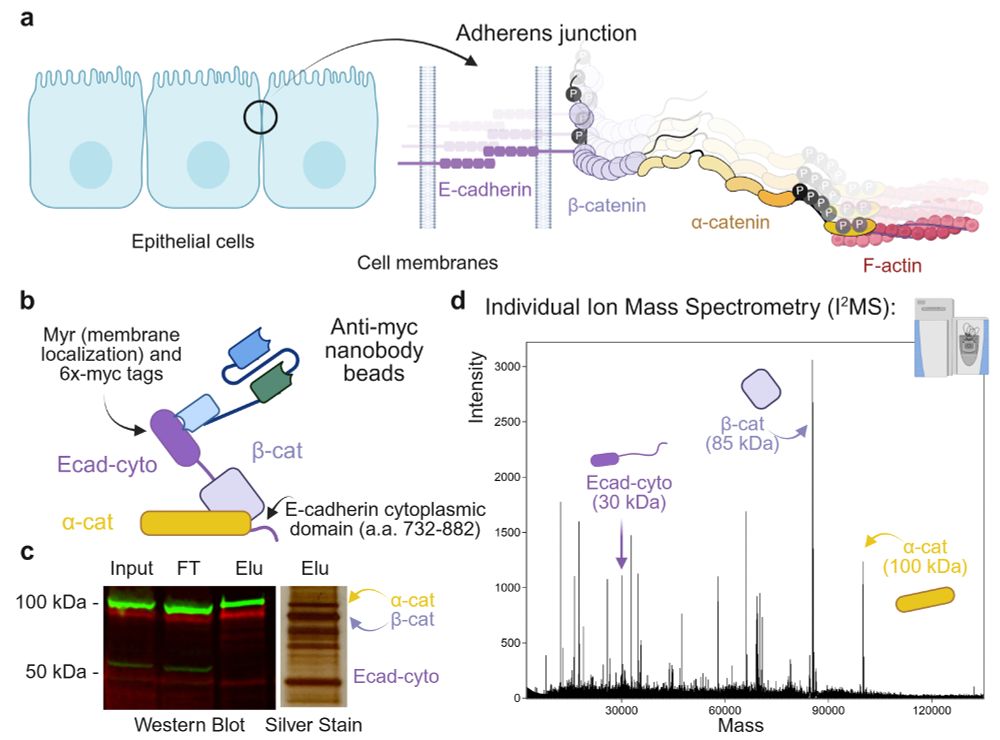
With Team @carajgottardi.bsky.social ‘s expertise in cell-cell adhesion, we purified cadherin-associated catenins and profiled their proteoforms using I2MS and found #phosphorylation to be the MAJOR modification. (3/7)
31.10.2025 20:00 — 👍 1 🔁 0 💬 1 📌 0

Team @nlkproteomics.bsky.social has developed individual ion mass spectrometry (I2MS) for proteoform analysis. We extend the current ~70 kDa mass limit beyond 100 kDa in this work. (2/7)
31.10.2025 20:00 — 👍 1 🔁 0 💬 1 📌 0

Heading to #HUPO2025! I’ll talk about our catenin phospho-code story and how top-down MS reveals how adhesion proteins respond to actomyosin force!
Thanks for the invite, #AFFINISEP!
Full story on bioRxiv 👉https://doi.org/10.1101/2025.09.06.674621
27.10.2025 15:44 — 👍 0 🔁 0 💬 0 📌 0

Neil Kelleher: The Human Proteoform Project Could Transform Medicine theanalyticalscienti...
---
#proteomics #prot-article
03.05.2025 19:20 — 👍 5 🔁 4 💬 1 📌 0

🚨 New in @naturemethods.bsky.social
#CHIMERYS has already transformed #proteomics since 2022, powering DDA, DIA & PRM analysis in a single workflow.
Wanna know how CHIMERYS works? 👉 www.nature.com/articles/s41...
#TeamMassSpec #NatureMethods #ASMS2025 🧪🔬
1/7
22.04.2025 11:10 — 👍 25 🔁 7 💬 3 📌 4

High-performance proteomics at any chromatographic flow rate
These data follow the expected trends as though taken from a textbook !
They illustrate clearly the trade-offs between high and low flow rates.
20.04.2025 20:43 — 👍 25 🔁 3 💬 2 📌 0
Sigh…
19.04.2025 20:16 — 👍 1 🔁 0 💬 0 📌 0
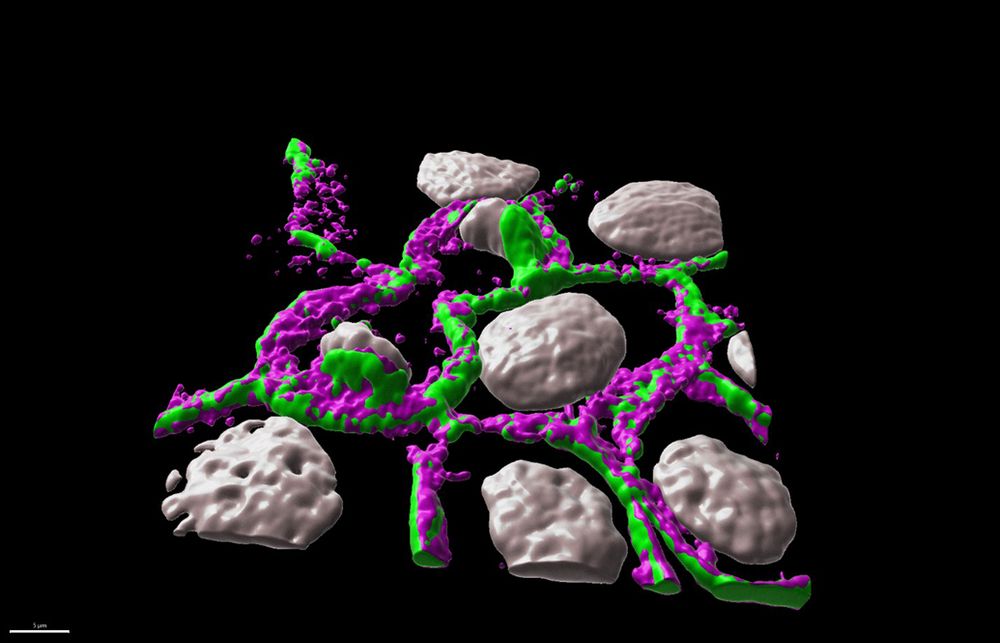
The phosphorylated form of α-catenin localizes to the apical portion of adherens junctions. The image shows 3D surface rendering using Imaris software of a confocal image of a dividing kidney epithelial cell line (Madin–Darby canine kidney or MDCK cells). Phosphorylated α-catenin is shown in magenta and adherens junctions are in green.
🧪Phuong Le, Jeanne Quinn, @jfhorner.bsky.social @carajgottardi.bsky.social & co show that the adhesion protein α-catenin has a key modification that allows dividing cells to stay better connected to their neighbours, helping the tissue stick together during mechanical stress. doi.org/10.1242/bio....
17.04.2025 13:39 — 👍 7 🔁 3 💬 1 📌 0

Looking forward to sharing my work at the Chicago Mass Spec Interest Group’s inaugural meeting!
05.04.2025 19:18 — 👍 2 🔁 0 💬 0 📌 0

It was a very fruitful #USHUPO2025 ! I was beyond thrilled to share our advances in catenin proteoform profiling and functional studies that will reveal the phospho-code in cell-cell adhesions. Many thanks to the conference organizers and participants!
27.02.2025 04:04 — 👍 1 🔁 0 💬 0 📌 0

Looking forward to attending the #USHUPO conference in Philly this weekend and sharing some early results in catenin proteoform profiling and our vision to build a story behind acto-myosin contractility regulation in cell-cell adhesion. Please join my talk next Wednesday and say hi!
17.02.2025 21:39 — 👍 1 🔁 0 💬 0 📌 0

Advancements in Global Phosphoproteomics Profiling: Overcoming Challenges in Sensitivity and Quantification analyticalsciencejou...
---
#proteomics #prot-paper
19.12.2024 10:20 — 👍 3 🔁 1 💬 0 📌 0

Emerging opportunities for intact and native protein analysis using chemical proteomics www.sciencedirect.co...
---
#proteomics #prot-paper
18.12.2024 16:00 — 👍 2 🔁 1 💬 0 📌 0
I love it when a place plays the Nutcracker during the holiday season, not All I Want For Christmas Is You on repeat 😝
18.12.2024 04:15 — 👍 0 🔁 0 💬 0 📌 0
Chemist/Physicist/Materials Scientist near Chicago.
Researching new ways to measure scent, better understand the cognition of taste and smell, find pollutants and toxics, measure quality of air and foods and add olfaction to robotic systems.
Product Manager @NautilusBio | Proteoform Aware | Views mine #Proteomics
Official account for National Taiwan University. Leading higher education and research in Asia since 1928. Follow for news, events, and achievements.
Wellcome Early Career Fellow @edinuni-irr.bsky.social @edinburgh-uni.bsky.social
| neutrophils single cell proteomics | road cyclist and aero bike enthusiast 🇨🇷🏴
Creator of the Immunological Proteome Resource @immpres.bsky.social
Systems & synthetic biologist, proteomics, bioinformatics; Professor of Molecular Biosciences @ UT Austin; Cofounder, Erisyon
#RNA #Virology & single-cell #Proteomics | Reader/Wellcome CDA fellow in
C4pr_liv
/livuniISMIB
| He/Him | Personal account - lab account: emmottlab@bsky.social
Web: emmottlab.org
Leading the Experimental Plant Systems Biology Lab at KU Leuven 🇧🇪. Previously at UAlberta 🇨🇦, ETH_Zurich🇨🇭, Imperial College London 🇬🇧, VIT University 🇮🇳.
Fmr Early Career Advisor @eLife, @biorxiv affiliate
he/him
mehta-lab.com
Group leader, Charlotte Group for Proteostasis Research | Associate Chair for Research in Biological Sciences at UNC Charlotte | Interested in the role of Hsp70 PTMs in yeast and mammalian cells | www.trumanlab.org
Scientist, still hanging around the UC Davis Genome Center. Really concerned with the state of things these days so you may see occasional political posts here or there
Associate professor at ETH Zurich, studying the cellular consequences of genetic variation. Affiliated with the Swiss Institute of Bioinformatics and a part of the LOOP Zurich.
Head of Data Science & Engineering at Talus Bio. I post about data science, BioML, AI drug discovery, Python, mass spec, proteomics, and silly things my kids do.
I also blog sometimes: https://willfondrie.com/post
Proteomics, humor, bbq, gravel riding, hiking, violin, gardening. Durham, NC
Focused (now) on analyzing whole 🩸 in sepsis. Pulmonary, Allergy and Critical Care Medicine, and Proteomics and Metabolomics Core Facility, Duke University
Professor of Proteomics. Currently at Newcastle University UK. he/him
Here in personal capacity.
Lab news and updates for the Emmott lab, RNA virology & proteomics at the Centre for Proteome Research, University of Liverpool, UK
Lab website: emmottlab.org
Research group at UNC-Chapel Hill under the direction of Dr. Erin S. Baker & focused on ion mobility-mass spectrometry research for biological and environmental systems.
Biologist and proteomics specialist at Max Perutz Labs Vienna.
Scientist studying molecules using mass spectrometry. Also coding sometimes.
ORCID: 0000-0002-5865-8994
I like big data and I cannot lie | Informatics/ML lead @ Belharra Tx, wet lab PhD @ Cravatt Lab
The number of people using expensive, black-box software for proteomics is bewildering.





























