Machine Learning-Driven Data Valuation for Optimizing High-Throughput Screening Pipelines
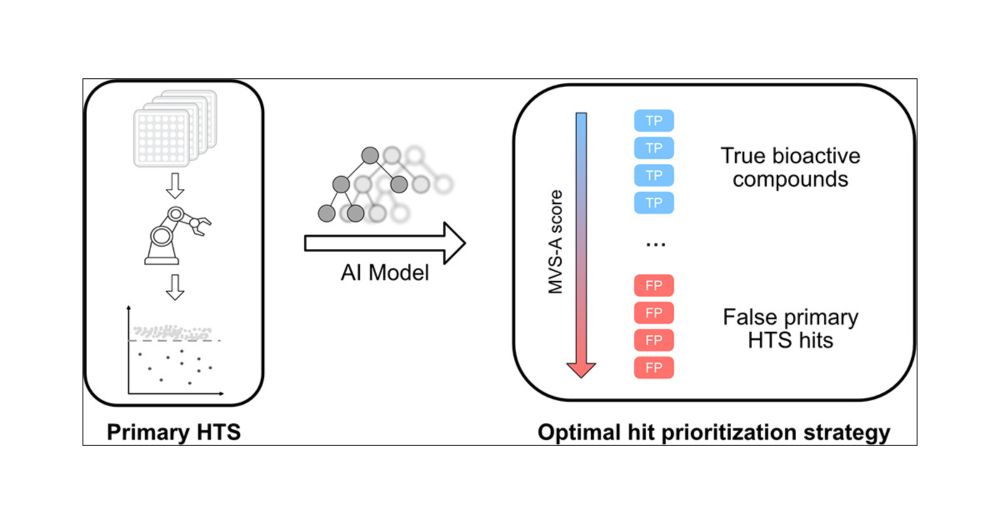
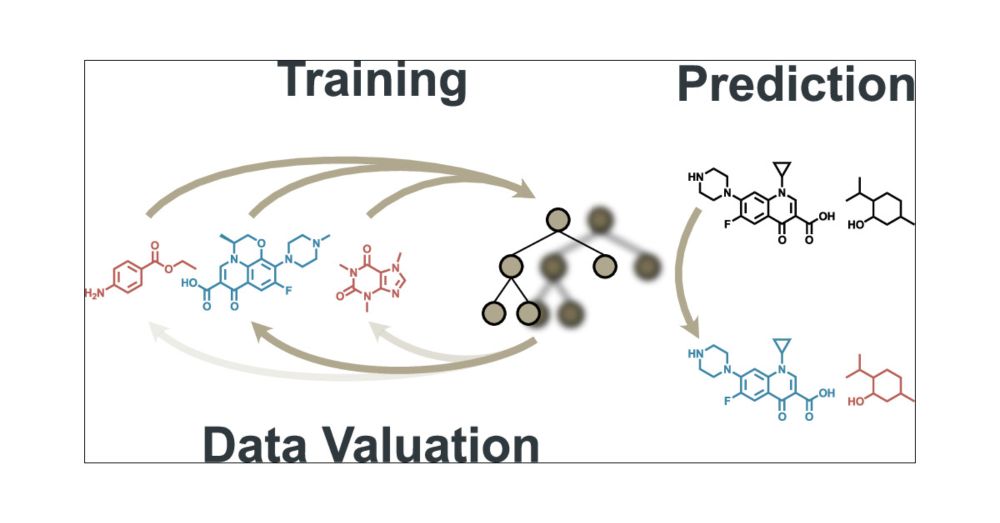
In the rapidly evolving field of drug discovery, high-throughput screening (HTS) is essential for identifying bioactive compounds. This study introduces a novel application of data valuation, a concept for evaluating the importance of data points based on their impact, to enhance drug discovery pipelines. Our approach improves active learning for compound library screening, robustly identifies true and false positives in HTS data, and identifies important inactive samples in an imbalanced HTS training, all while accounting for computational efficiency. We demonstrate that importance-based methods enable more effective batch screening, reducing the need for extensive HTS. Machine learning models accurately differentiate true biological activity from assay artifacts, streamlining the drug discovery process. Additionally, importance undersampling aids in HTS data set balancing, improving machine learning performance without omitting crucial inactive samples. These advancements could significantly enhance the efficiency and accuracy of drug development.
Our new paper leverages data valuation concepts, improving early stage drug discovery processess - enhancing large library screening via active learning and distinguishing true from false positives according to sample importances for faster, efficient hit identification.
pubs.acs.org/doi/full/10....
05.02.2025 12:38 — 👍 0 🔁 0 💬 0 📌 0

Chemiker Prof. Stephan A. Sieber erhält Inhoffen-Medaille 2024
Für seine Forschung zu neuen Medikamenten gegen multiresistente Bakterien zeichnen ...
We were grateful to receive last year's Inhoffen-Medaille 2024 by the Förderverein des HZI and TU Braunschweig on our drug developing efforts against multidrug resistant bacteria. We want to thank everyone involved for making this possible!
magazin.tu-braunschweig.de/m-post/chemi...
21.01.2025 15:23 — 👍 5 🔁 1 💬 0 📌 0
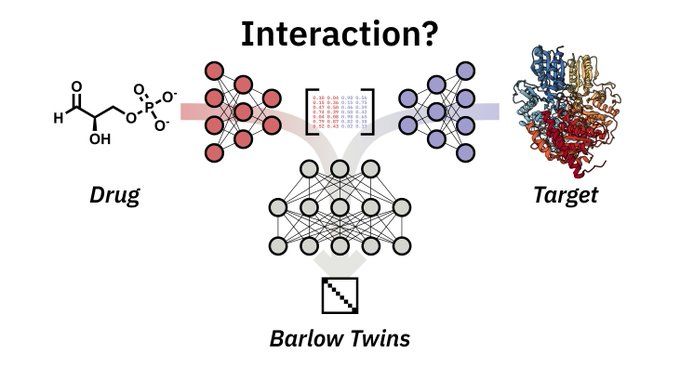
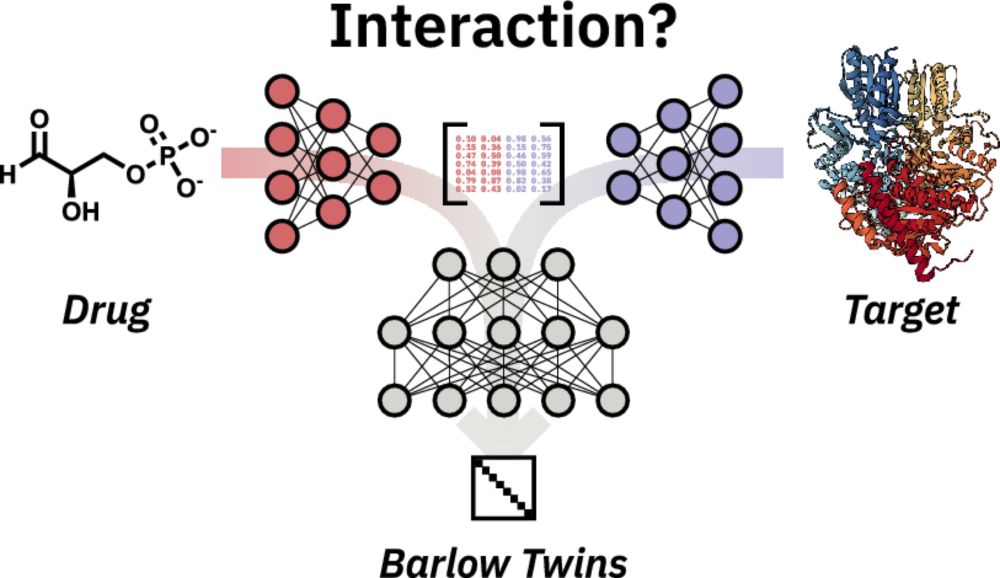
Exciting news in drug discovery! Our "new" paper (back in September 2024) introduces BarlowDTI, a cutting-edge method using the Barlow Twins architecture to predict drug-target interactions with state-of-the-art performance. Check it out: arxiv.org/abs/2408.00040
21.01.2025 15:21 — 👍 2 🔁 0 💬 0 📌 0
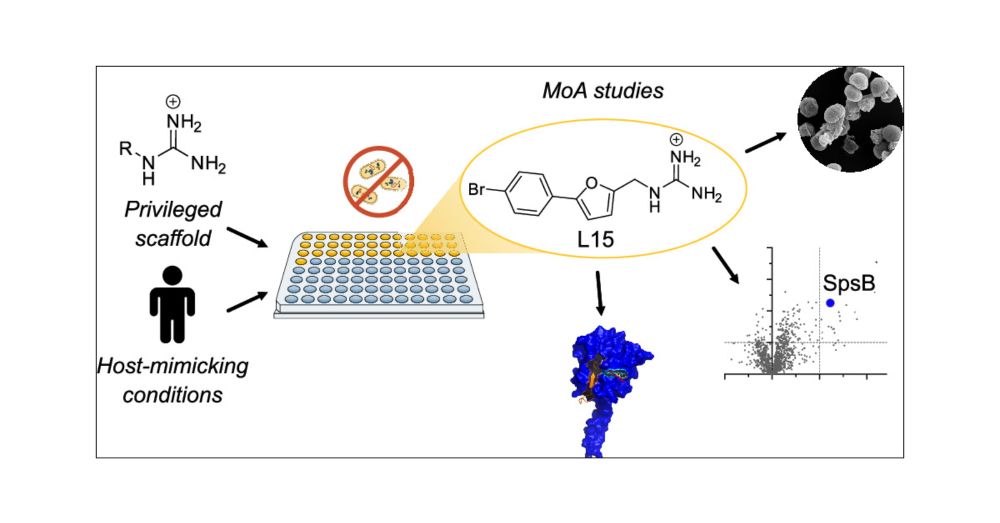
Screening Privileged Alkyl Guanidinium Motifs under Host-Mimicking Conditions Reveals a Novel Antibiotic with an Unconventional Mode of Action
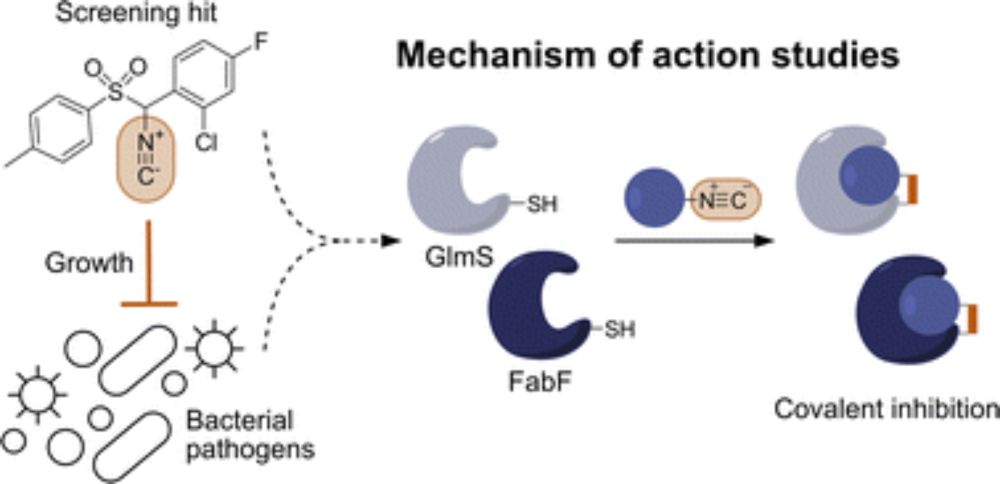
Screening large molecule libraries against pathogenic bacteria is often challenged by a low hit rate due to limited uptake, underrepresentation of antibiotic structural motifs, and assays that do not resemble the infection conditions. To address these limitations, we present a screen of a focused library of alkyl guanidinium compounds, a structural motif associated with antibiotic activity and enhanced uptake, under host-mimicking infection conditions against a panel of disease-associated bacteria. Several hit molecules were identified with activities against Gram-positive and Gram-negative bacteria, highlighting the fidelity of the general concept. We selected one compound (L15) for in-depth mode of action studies that exhibited bactericidal activity against methicillin-resistant Staphylococcus aureus USA300 with a minimum inhibitory concentration of 1.5 μM. Structure-activity relationship studies confirmed the necessity of the guanidinium motif for antibiotic activity. The mode of action was investigated using affinity-based protein profiling with an L15 probe and identified the signal peptidase IB (SpsB) as the most promising hit. Validation by activity assays, binding site identification, docking, and molecular dynamics simulations demonstrated SpsB activation by L15, a recently described mechanism leading to the dysregulation of protein secretion and cell death. Overall, this study highlights the need for unconventional screening strategies to identify novel antibiotics.
Another cool story of us is published in JACSAu! We screened alkyl guanidinium scaffolds under host-mimicking conditions to identify novel antibiotics and elucidated an unconventional mode of action of our most promising hit compound L15.
pubs.acs.org/doi/10.1021/...
21.01.2025 15:19 — 👍 1 🔁 0 💬 0 📌 0
Scientist, #MachineLearning and #AI for Moleculear Sciences. Scuba Diver. Loves @cecclementi.bsky.social
Director of the Institute of AI for Health @ Helmholtz Munich
Presidential Associate Professor @upenn.edu - using AI to reimagine antibiotic discovery and peptide design. Previously @MIT, @UBC
🔗 https://delafuentelab.seas.upenn.edu
Computational biologist @HelmholtzMunich, prof @TU_Muenchen & associate PI @sangerinstitute. Dad of 4 and mountain lover. Department news, see @CompHealthMuc
Principal Researcher at Microsoft Research AI for Science, ML for Drug Discovery, ex Google AI & tensorflight
Liebig Fellow Group Leader at Ruhr University Bochum, Laboratory for Medicinal Inorganic Chemistry
https://www.kargesgroup.ruhr-uni-bochum.de/
Structural biologist, ubiquitin enthusiast, Chemical Biology, amateur photographer. Views are my own.
News from Jennifer Doudna's lab at UC Berkeley, Innovative Genomics Institute. Tweets from lab members and not Jennifer Doudna unless signed JD. Tweets represent personal views only.
doudnalab.org
Chemist, Dad, Group Leader, Associate Professor. Excited about novel chemical modalities in drug discovery
Scientist 🌍collaboration advocate🤝#interdisciplinary #research for smart & #sustainable (nano)technologies🧪⚡️💡as Associate #Prof at Uni Turku 🇫🇮 & Adolphe Merkle Institute🇨🇭invested in #Science4Policy&Diplomacy while 😷 m⌬m 🌐 www.jovanamilic.com
Bio-organic synthetic lab with focus on the synthesis of ADP-ribose related biopolymers.
chemical biology enthusiast & strategist; I make impossible things possible - all before breakfast!
Assistant Professor at Caltech | Chemical Biologist
morsteinlab.caltech.edu
Assistant professor Chemical Genetics @ Leiden University. Interests: Drug discovery, Chemical biology, Target discovery and validation, Gene editing.
University of Arizona. Studying all things PTM and metabolism. Galliganlab.com
Group specialized in the synthesis of foldamers based on aromatic oligoamides & biological applications 🧬
| LMU Munich, Germany | huc.cup.uni-muenchen.de
#ChemBio #Supra
(account run by postdoc @lggchem.eu)
Passionate scientist, runner, basket. Group leader at #IRBBarcelona | Targeted protein degradation | Chemical biology