I'm excited to share our pre-print about a new variant benchmarking tool we've been working on for the past few months!
Aardvark: Sifting through differences in a mound of variants
GitHub: github.com/PacificBiosc...
Some highlights in this thread:
1/N
06.10.2025 20:07 — 👍 29 🔁 15 💬 1 📌 0
🦒Long read giraffe is out!🦒
Mapping long reads to pangenome graphs is ~10x faster than with GraphAligner, with veeery slightly better mapping accuracy, short variant calling, and SV genotyping than GraphAligner or Minimap2
02.10.2025 06:28 — 👍 41 🔁 22 💬 1 📌 0

Do you know ~60% of human SVs fall in ~1% of GRCh38? See our new preprint: arxiv.org/abs/2509.23057 and the companion blog post on how we started this project and longdust: lh3.github.io/2025/09/29/o.... Work with Alvin Qin
30.09.2025 02:19 — 👍 75 🔁 27 💬 0 📌 0

I'm hiring a computational biologist interested in complex trait genetics using deep learning approaches. Reach out to me, if interested.
12.09.2025 19:00 — 👍 36 🔁 46 💬 1 📌 0
Now published in GigaScience with minor improvements: academic.oup.com/gigascience/...
* Download: zenodo.org/records/1490...
* More info: github.com/lh3/panmask
04.09.2025 16:44 — 👍 30 🔁 10 💬 1 📌 1

GitHub - jltsiren/gbz-base: Prototype for an immutable pangenome graph in SQLite
Prototype for an immutable pangenome graph in SQLite - jltsiren/gbz-base
GBZ-base has been a side project for me for a couple of years. It's basically a GBZ graph stored in SQLite instead of a custom file format. You can convert a GBZ graph to GBZ-base quickly and then extract subgraphs around nodes / reference positions on a laptop. 1/n
28.08.2025 00:49 — 👍 5 🔁 2 💬 2 📌 0

Interesting job listings 👀 astromech.com
www.sec.gov/Archives/edg...
15.08.2025 17:19 — 👍 1 🔁 2 💬 0 📌 0

Postdoctoral Scholar – Genomics, Aging, Somatic Mutation, Structural Variation, Evolution , Cancer – Integrative Biology
University of California, Berkeley is hiring. Apply now!
The Sudmant lab at UC Berkeley is seeking a postdoc to work on a fully funded NIH project to understand differences in DNA repair and somatic mutation across the primate tree of life. Please spread widely to those who may be interested aprecruit.berkeley.edu/JPF05052
13.08.2025 04:22 — 👍 54 🔁 55 💬 0 📌 1
This sounds true to me
05.08.2025 10:34 — 👍 33 🔁 7 💬 2 📌 0

Beeswarm plot of the prediction error across different methods of double perturbations showing that all methods (scGPT, scFoundation, UCE, scBERT, Geneformer, GEARS, and CPA) perform worse than the additive baseline.

Line plot of the true positive rate against the false discovery proportion showing that none of the methods is better at finding non additive interactions than simply predicting no change.
Our paper benchmarking foundation models for perturbation effect prediction is finally published 🎉🥳🎉
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
04.08.2025 13:52 — 👍 128 🔁 57 💬 2 📌 6
Technical blogging for growth and learning
Writing a technical blog improves your writing, forces you to learn new things, helps others and yourself, and helps your career. This post: why blog, what to blog about, how to start, and using AI.
Writing a technical blog improves your writing, forces you to learn new things, helps others and yourself, and helps your career. In this essay: (1) why blog, (2) what to blog about, (3) how to get started, and (4) using AI. https://doi.org/10.59350/bqnfd-7p249 🧪
25.01.2025 16:00 — 👍 54 🔁 11 💬 2 📌 2
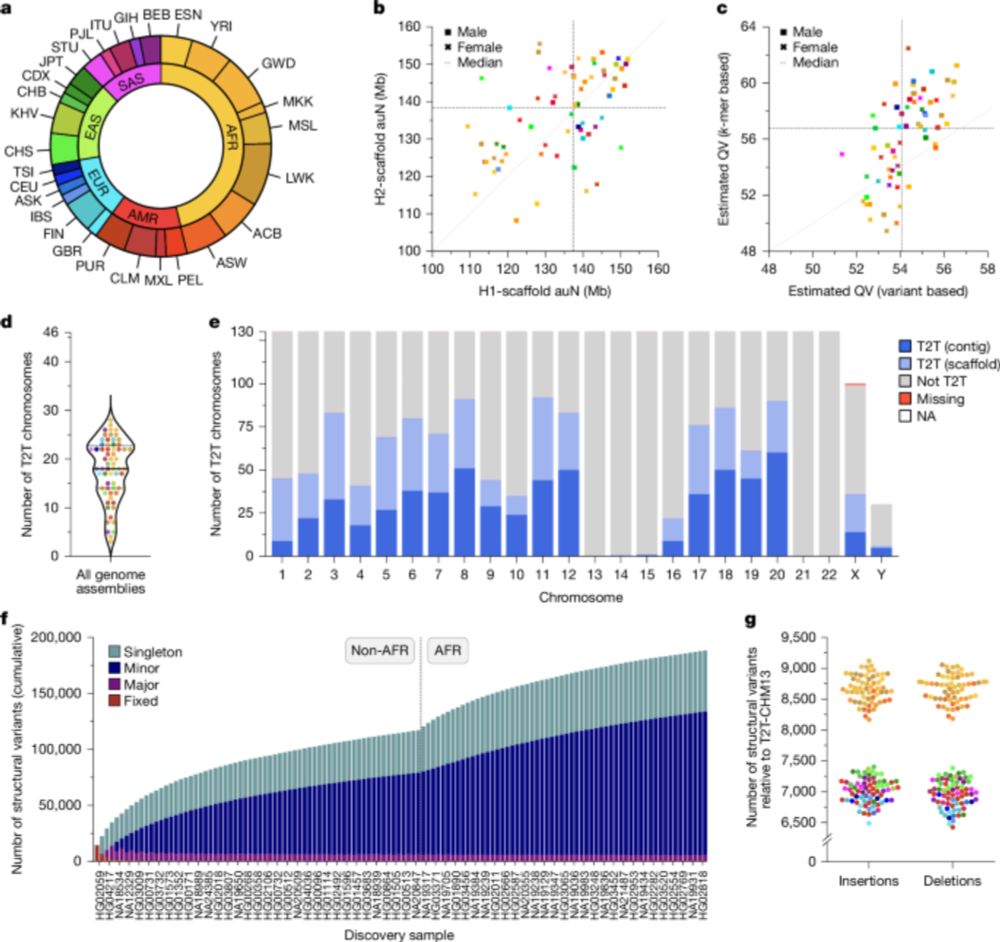
65 Genomes Expand Our Picture Of Human Genetics
Researchers closely examined the genomes of 65 individuals to paint a more complex, and more complete, picture of human genetic diversity.
Complete genomes alert! @glennislogsdon.bsky.social, @christinebeck.bsky.social, and I were on @scifri.bsky.social today talking about "Complex genetic variation in nearly complete human genomes"
📄 www.nature.com/articles/s41...
📻 www.sciencefriday.com/segments/65-...
01.08.2025 20:49 — 👍 50 🔁 16 💬 1 📌 2

Preprint on "Finding easy regions for short-read variant calling from pangenome data": arxiv.org/abs/2507.03718
08.07.2025 02:14 — 👍 31 🔁 13 💬 0 📌 1
This a really exciting leap forward for genomic sequence to activity gene regulation models. It is a genuine improvement over pretty much all SOTA models spanning a wide range of regulatory, transcriptional and post-transcriptional processes. 1/
25.06.2025 16:18 — 👍 72 🔁 20 💬 2 📌 2
Excited to launch our AlphaGenome API goo.gle/3ZPUeFX along with the preprint goo.gle/45AkUyc describing and evaluating our latest DNA sequence model powering the API. Looking forward to seeing how scientists use it! @googledeepmind
25.06.2025 14:29 — 👍 219 🔁 82 💬 5 📌 10

An assessment of DNA language models concludes:
◼️ They do not offer compelling gains over baseline models
Their performance is inconsistent and requires much more compute.
arxiv.org/abs/2412.05430
23.06.2025 20:21 — 👍 52 🔁 19 💬 1 📌 3

Preprint on "Improving spliced alignment by modeling splice sites with deep learning". It describes minisplice for modeling splice signals. Minimap2 and miniprot now optionally use the predicted scores to improve spliced alignment.
arxiv.org/abs/2506.12986
17.06.2025 01:48 — 👍 109 🔁 54 💬 0 📌 1
A blueprint for the batmobile.
Your genome is not a blueprint. A thread about misleading metaphors in science communication. 🧬🧪 1/n
14.06.2025 15:49 — 👍 199 🔁 66 💬 9 📌 9
People always stop me in the street to ask: "Yoav, where are the disease-associated eQLTs? We found a lot in GTEx but we can't find anymore. Do you know where they are?"
(For the record, no one has ever asked me this, but it is a really good question!)
I think we know where they are.
10.06.2025 14:20 — 👍 58 🔁 14 💬 3 📌 2

Standard methods are equivalent to a flashlight, looking at each gene independently. We combine signals from multiple genes, turning a floodlight onto the genome.
Excited to share my first PhD paper in the @sbmontgom.bsky.social lab with @tamigj.bsky.social (www.biorxiv.org/content/10.1...)! Standard QTL methods treat each gene independently. But what if a single variant regulates multiple nearby genes at once - what we call “allelic proxitropy”? 🧵 ⬇️
08.06.2025 17:38 — 👍 91 🔁 33 💬 6 📌 4
Very excited to share I’ll be starting my own group at University of Utah in the Department of Human Genetics in the new year! Reach out if you are interested! vollgerlab.com
05.06.2025 14:15 — 👍 65 🔁 12 💬 9 📌 4
Neng Huang developed longcallR for joint SNP calling and phasing from long RNA-seq reads, AND for identifying allele-specific splicing/junctions (ASJ). Although ASJs of statistical significance are rare, a large fraction involve unannotated junctions. In Rust!
30.05.2025 14:54 — 👍 16 🔁 7 💬 0 📌 0
The human pangenome continues to grow and improve! Release 2 is here! Click through for the details, but this is a pretty amazing dataset including not just the phased assemblies, but PacBio HiFi, ONT Ultralong, Dovetail/Illumina Hi-C, PacBio Kinnex, and Illumina WGS for all samples
12.05.2025 13:52 — 👍 82 🔁 36 💬 0 📌 0
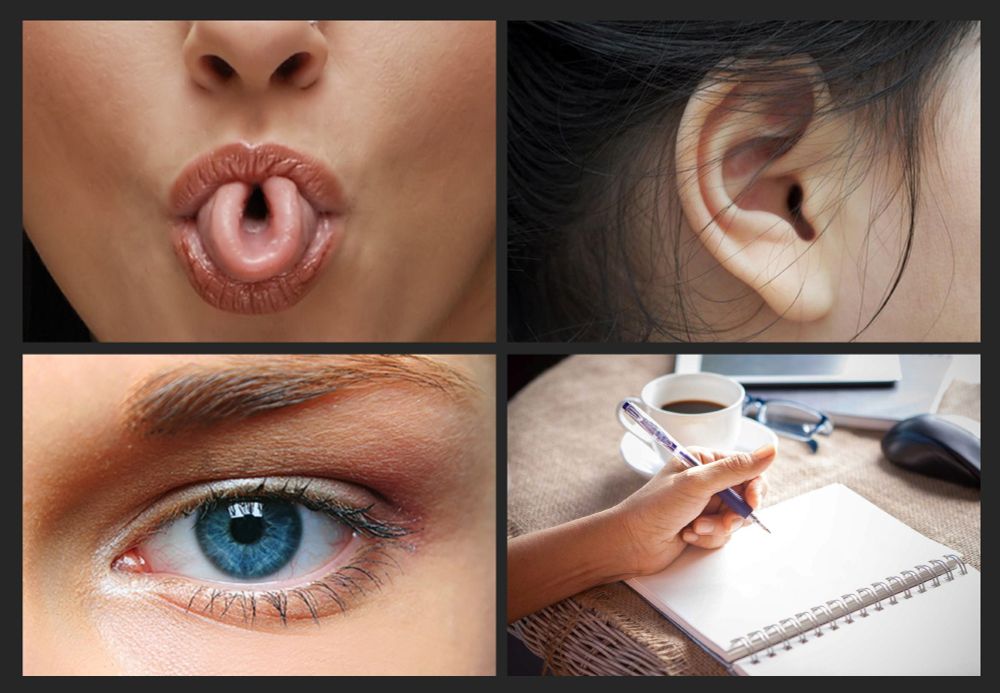
Four images to illustrate some prominent single-gene myths. Top left shows a photograph of a person deftly rolling their tongue into a U-shape. Top right shows a photograph of a person’s ear, highlighting the shape and features of the earlobe and cartilage. Bottom left shows a close-up photograph of a person’s eye, with a vivid blue colouration. Bottom right shows a photograph of a person poised to write with their left hand on the blank white page of a spiral-bound notebook.
Remember when you first learned about genetics at school? All those fascinating examples of human traits that are each apparently determined by just a single gene? Time to check in on some of your favourites to see how they’re doing. 🧬🧵🧪 1/n
02.05.2025 14:50 — 👍 1265 🔁 595 💬 51 📌 83
Staff scientist at @PacBio; formerly @hudsonalpha; avid gamer; opinions are my own
Product manager @PacBio / RNA / bioinformatics / aerialist & pole dancer / opinions are my own and do not reflect views of my employer
Genomics, Bioinformatics.
github.com/tobiasrausch
We are a world leader in genomics research. We apply and explore genomic technologies to advance the understanding of biology and improve health 🧬
https://www.sanger.ac.uk/
Principal Investigator at Boston Children's Hospital | Statistical Genetics
Sudmantlab.org - Assistant professor Berkeley
Postdoc at MD Anderson: molecular epi + statistical genomics of human development & cancer | PhD, Penn State: functional genomics in social insects.
Opinions are my own.
https://seantbresnahan.com
https://bhattacharya-lab.com
We’re working to enable faster and deeper genomic research, to bring genomic healthcare to all who need it.
Associate Professor of Genetics and Genome Sciences at UConn Health and The Jackson Laboratory
Transposons, Structural Variation, Genomics, Gardening, Loud Music, Mom
The NHGRI-EBI Catalog of genome-wide association studies (GWAS)
www.ebi.ac.uk/gwas
Submit your #gwas data on http://bit.ly/38rNSjx
#OpenAccess #FAIRprinciples
MBBS, MD, PhD | GWAS storyteller | Scientist at Regeneron | Human genetics & drug discovery in Neuroscience & Psychiatry
official Bluesky account (check username👆)
Bugs, feature requests, feedback: support@bsky.app
Bioinformatics and Computational Genomics at @HHU.de. Algorithms for pangenomes, structural variation, genome assembly, haplotype phasing, etc.
The goal of the Canadian BioGenome Project is to produce high-quality reference genomes 🧬 for all Canadian species 🌎
Sequencing Canada's Biodiversity 🌿🦋🐍🧬🐢🦌🐸🌳🐿🐙🦈🦦
Learn more: https://linktr.ee/canadianbiogenome
The Earth BioGenome Project (EBP), a moonshot for biology, aims to sequence, catalog, and characterize the genomes of all of Earth's eukaryotic biodiversity over a period of ten years.
🌲Keep up with all EBP updates: https://linktr.ee/earthbiogenomeproject
Research scientist at @GoogleDeepMind passionate about AI, genomics and biology.
Associate Professor of CS @ University of Maryland. Proud Rust advocate! I ♥ science & compiled, statically-typed programming languages! Views are my own. Tech stack: https://github.com/rob-p/tech-stack.