Loss of SUMOylation drives aberrant PRC1 clustering and 3D genome rewiring independent of H3K27me3 https://www.biorxiv.org/content/10.64898/2026.02.05.704038v1
07.02.2026 06:33 — 👍 6 🔁 3 💬 0 📌 0

Epigenetics Update - DOT1L provides transcriptional memory through PRC1.1 antagonism go.nature.com/4qh5aXB
Chen Davidovich and Omer Gilan (Monash University) reporting in Nat Cell Biol
#Epigenetics #DOT1L #PolyComb
---
Gain deeper insights into gene regulation; epigenometech.com
06.02.2026 12:50 — 👍 8 🔁 2 💬 0 📌 0

Science update from my postdoc Peiyuan Chai - his work “glycoRNA complexed with heparan sulfate regulates VEGF-A signaling” is now published @nature.com uncovering a new layer or glycoRNA-regulation of growth factor mediated control physiological processes rdcu.be/e1bBX
28.01.2026 17:45 — 👍 78 🔁 28 💬 6 📌 3

Two alternative isoforms of PRC2 accessory subunit AEBP2 modulate developmental Polycomb functions in opposite ways - with broadly-expressed AEBP2L acting as an intrinsic inhibitor in somatic cells
@adrianbracken.bsky.social @davidovichlab.bsky.social and colleagues
www.embopress.org/doi/full/10....
04.11.2025 12:51 — 👍 13 🔁 5 💬 0 📌 1
YouTube video by KeystoneSymposia
KSQA: Dr Karim-Jean Armache / Dr. Cigall Kadoch (Epigenetics and Gene Regulation)
@ckadoch.bsky.social and I are excited to welcome you to Geneva for the 2026 Keystone Symposium on Epigenetics and Gene Regulation in Health and Disease — short talk and poster slots are still open. Don’t miss the deadlines.
Video: youtu.be/sLfyuQuH8F0
03.11.2025 14:43 — 👍 14 🔁 7 💬 0 📌 0
2/ Done in collaboration with the lab of @adrianbracken.bsky.social, who wrote a great 🧵 explaining the work. bsky.app/profile/adri...
31.10.2025 22:42 — 👍 0 🔁 0 💬 0 📌 0
1/ 🚀 AEBP2 isn’t what we thought.
You were told that AEBP2 promotes PRC2 activity on chromatin.
We found the opposite: the most prevalent AEBP2 isoform inhibits PRC2 activity.
👉 surl.li/cgwqcq
A thread 🧵
31.10.2025 10:53 — 👍 40 🔁 16 💬 6 📌 2

Abstract extension flyer. Deadline extended to Friday 31st October
📣 Abstract Deadline Extended!
Due to popular demand, we’ve extended the submission deadline — giving you a few extra days to finalise your abstract.
Don’t miss out on being part of Lorne’s legacy!
👉 Submit today: www.lorneproteins.org
#LorneProteins #DeadlineExtended #ProteinScience
26.10.2025 22:59 — 👍 5 🔁 3 💬 0 📌 1

Screenshot of tweet from the ARC saying they will announce DP26 outcomes on Tuesday 28th Oct, and LP25r1 outcomes on Wednesday 29th.
ARC says Discovery Projects outcomes will be tomorrow (Tuesday). Linkage Projects (2025, round 1) on Wed. Over past ~year, it's often been at about 11am (Canberra).
My bot will pick up the change to RMS & post immediately.
ARC should email outcomes to lead CIs, but might take an 1hr or so for DPs
26.10.2025 23:17 — 👍 53 🔁 27 💬 1 📌 3

EEE meeting is BACK! Early Embryogenesis & Epigenetics conference in Berlin 02/2026.
Checkout great program and over 12 slots for (not so) short talks for submitted abstracts!. Early registration now open -
w.molgen.mpg.de/embryo2026
28.09.2025 00:00 — 👍 29 🔁 12 💬 0 📌 1

Genes and Gin: A Synthetic Biology Social 🎉
Join us for a fun night of Genes and Gin - learn to play the synthetic card game "Remediate!", created by a group of researchers within our Centre.
Make sure you register for catering:
www.eventbrite.com.au/e/genes-and-...
See you on the rooftop!
18.09.2025 00:58 — 👍 2 🔁 1 💬 0 📌 0
Many of the most complex and useful functions in biology emerge at the scale of whole genomes.
Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
17.09.2025 15:03 — 👍 49 🔁 20 💬 3 📌 4

The PRC2-associated factor EPOP is required for Hox gene regulation during axial development in mice
The Polycomb repressive complex 2 (PRC2) is an essential modulator of gene repression. We previously reported that, in mouse embryonic stem cells, PRC…
Finally out on @devbiol.bsky.social: we find that Epop mutant mice have near-perfect Polycomb phenotype (orderly posterior shift of vertebral identity🩻) linked to accelerated HOX clock⏰.
Bonus: Epop has maternal effect!
Study co-led by the the labs of Bernie Payer and @lucianodicroce.bsky.social
25.08.2025 19:44 — 👍 22 🔁 3 💬 2 📌 3

We are excited to share our new preprint demonstrating that nucleic acid interactions with SUZ12 constrain PRC2 activity, establishing a kinetic buffer essential for targeted gene silencing and revealing vulnerabilities in diffuse midline gliomas.
www.biorxiv.org/content/10.1...
23.07.2025 23:38 — 👍 48 🔁 19 💬 1 📌 0
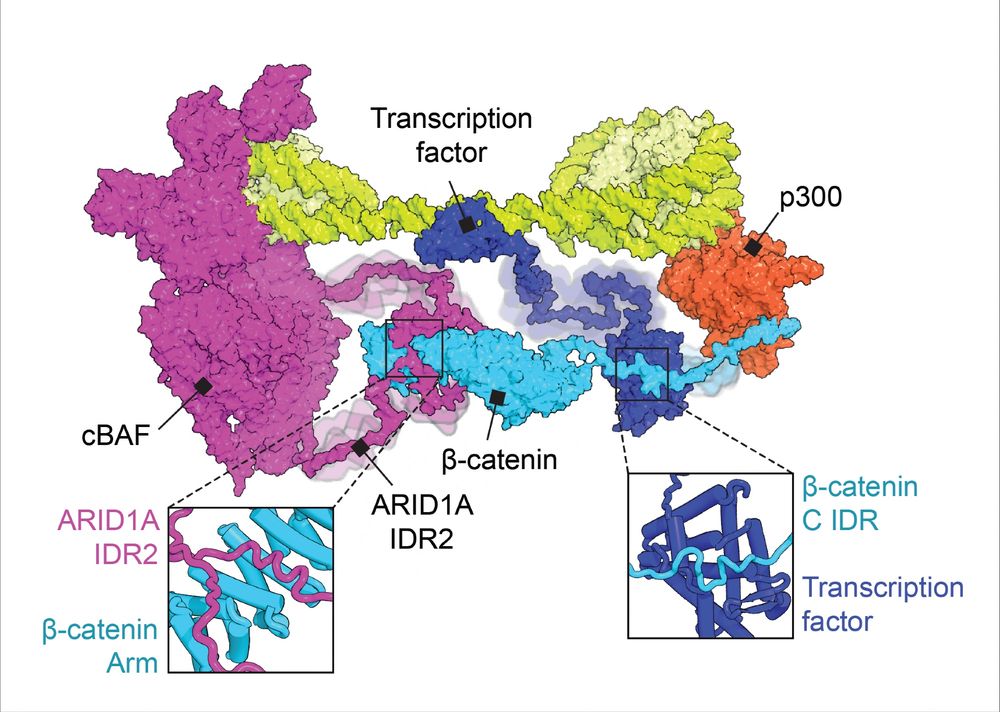
How do chromatin remodelers use #IDRs to find TF binding partners? In our new Molecular Cell paper, we show that β-catenin is an adaptor that links SWI/SNF (cBAF) subunit ARID1A with binding partners via IDR-domain interactions.
www.cell.com/molecular-ce...
22.07.2025 18:42 — 👍 89 🔁 34 💬 11 📌 4
Australian Cell Cycle, DNA repair and Telomere Meeting
October 20-22, 2025
Register for the Australian Cell Cycle, DNA Repair & Telomere Meeting in beautiful Melbourne October 19-22, 2025. Plenty of slots for selected abstracts and posters. High praise last time from Piotr Sicinski: "en par with a Gordon conference for science and interaction"
www.australiancellcycle.org
22.07.2025 10:42 — 👍 8 🔁 4 💬 0 📌 1
Happy to announce a new preprint from my lab looking in to the establishment of polycomb domains in early fly development and contributions from pioneer factors Zelda and GAGA-factor.
03.07.2025 21:44 — 👍 20 🔁 7 💬 0 📌 0
✨Excited to share my first postdoc paper on chromatin remodeler, MORC2 ✨
Using #cryoEM & #singlemolecule imaging, we reveal how MORC2 compacts DNA 🔬🧬
Thanks to all collaborators from @wehi-research.bsky.social @ccemmp-outreach.bsky.social Uni Melb, Monash Uni, Seoul National Uni and Texas Uni!
02.07.2025 06:19 — 👍 13 🔁 5 💬 0 📌 0
Postdoc in the Gahan Lab at the CCB in Galway, Ireland 🍀 Interested in the origin of animals from the point of view of their closest living relatives 🧫
Developmental biologist | Professor at Indiana University Department of Biology | eye development, pattern formation, gene regulation
Group leader at @cbgpmadrid.bsky.social working on #epigenetics and #genomics in Arabidopsis and Brassica crops. Tweets science & related stuff in english and español.
Bioinformatics, protein modeling, cryoEM, drug screening, function prediction. Daisuke Kihara, professor of Biol/CS, Purdue U. https://kiharalab.org/ YouTube: http://alturl.com/gxvah
Assistant Professor 👩🏼💻 at Erasmus Medical Center Rotterdam researching genetics & gene regulation 🧬🖥 in the context of disease 🥼💉📊
Functional Genomics, Rare Diseases and Cardiovascular Disease @ Exeter University
ORCID 0000-0001-8074-6299
Enhancer biology at Rada-Iglesias lab
Biochemist fascinated by visualizing biological processes with the finest spatiotemporal detail; Understand the human immune system to guide novel approaches for combating viral infections and cancer; Postdoc at Cissé Lab / MPI-IE
New PI @mpi_ie, interested epigenetics, development, homologous chromosome interactions and their function (she/her)
Fosters productive collaborations, rigorous debate, and enduring friendships between RNA scientists across Colorado. Fueled by 🍕, 🍺, and 🧬 since 1986.
🇲🇹 🇬🇧 Exploring the weird and wonderful world of #redalgae www.borglab.org
Research Associate in the Ferguson-Smith Lab @ Cambridge
PhD in the Bickmore Lab @ Edinburgh
🧬 https://orcid.org/0000-0001-7025-7002 🧬
Retired researcher in Developmental Genetics, Evo-Devo. Drosophila. Polycomb Group. (PRC genes. Finder of "polyhomeotic"), Nuclear transplantation in the egg. Cell mosaics and chimeras. Epigenesis in Drosophila. CSIC --> CNRS. Hobby: mosaics.
We are the Ting Wu Lab @ HMS. Our laboratory studies how chromosome behavior and positioning influence genome function, with implications for gene regulation, genome stability, and disease.
Managed by WuLab members
https://www.transvection.org/home
Scientist and medical doctor. Biology AI/ML methods, gene regulation, DNA sequence models, single cells. Doing a PhD in computational biology at @molgen.mpg.de.
Assistant Professor of Nutritional Sciences UW-Madison studying the genomics of inflammatory disease and vitamin D metabolism.
All views and opinions are my own and not reflective of the University of Wisconsin-Madison. @UWmadison
Postdoc in Claudia Bank’s group at the University of Bern. Promoters, emergence, fitness landscapes, dogs 🐶 and triathlon 🏊 🚲 🏃. He/him 🏳🌈.
timothyfuqua.com
Faculty in Genome Sciences at the University of Washington focusing on gene regulation tech dev, especially in plants.
PhD student in the San Roman lab in the Molecular Genetics and Microbiology program at Duke Univerity🧬 University of Vermont ‘23 ⛰️


























