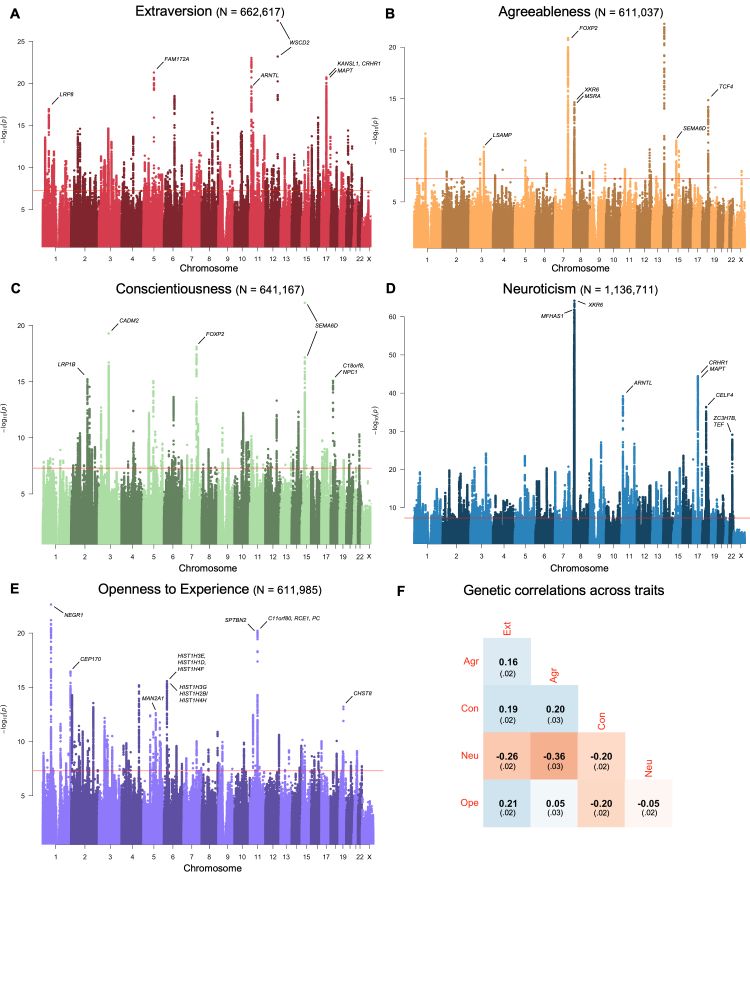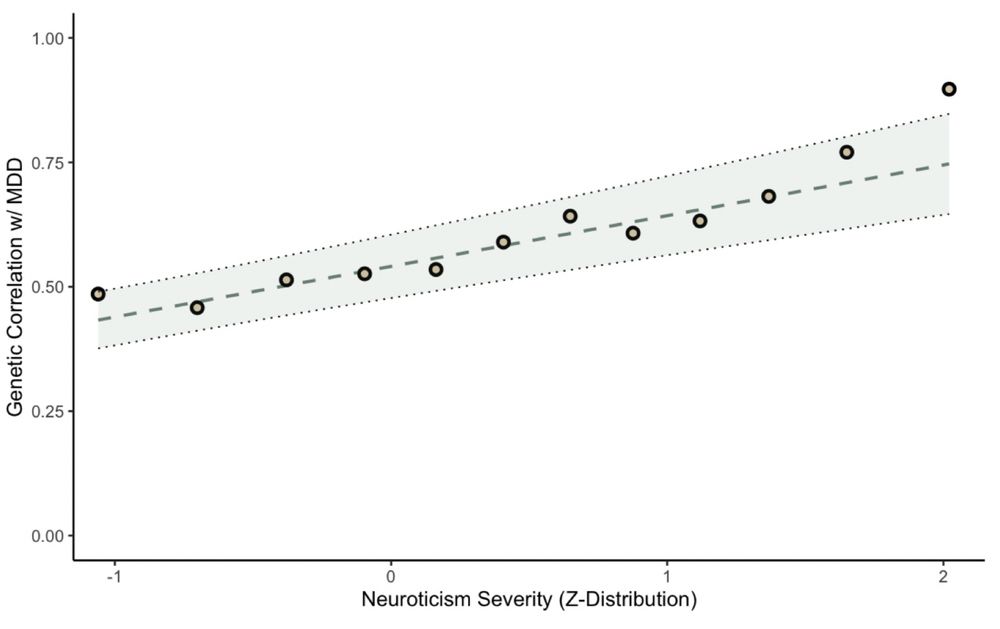
Genetic correlations between MDD and each neuroticism cut point. The line of best fit is shown, based
on a linear regression model, with parameters estimated using generalized least squares (GLS).
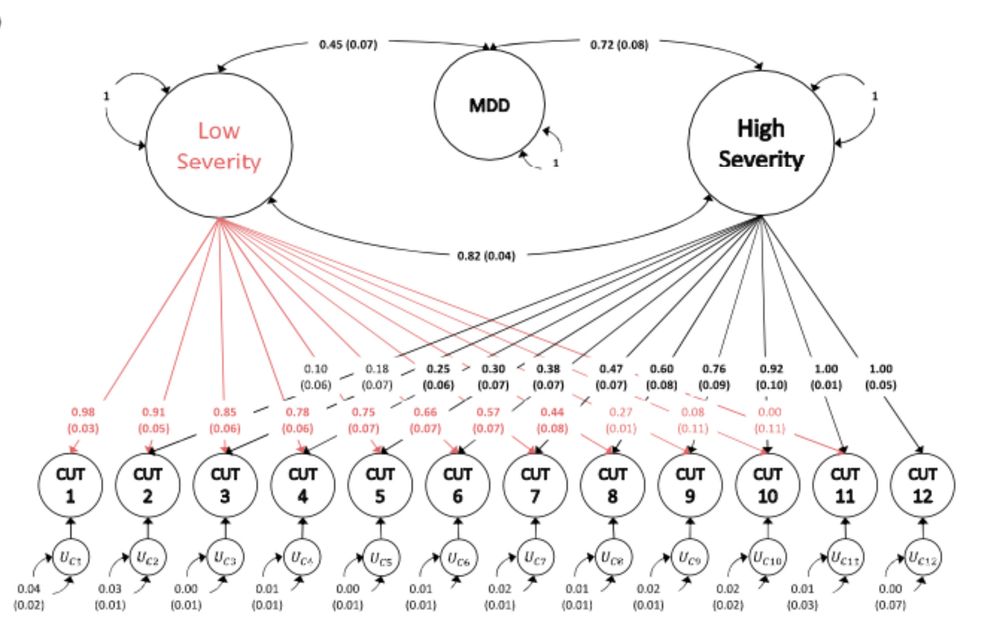
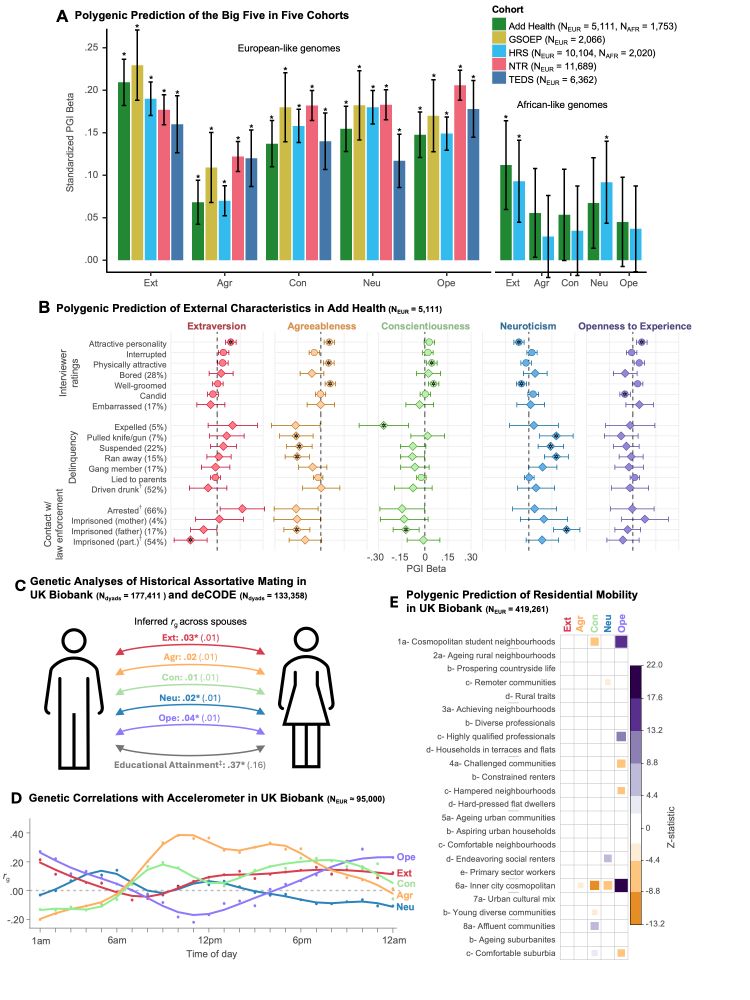
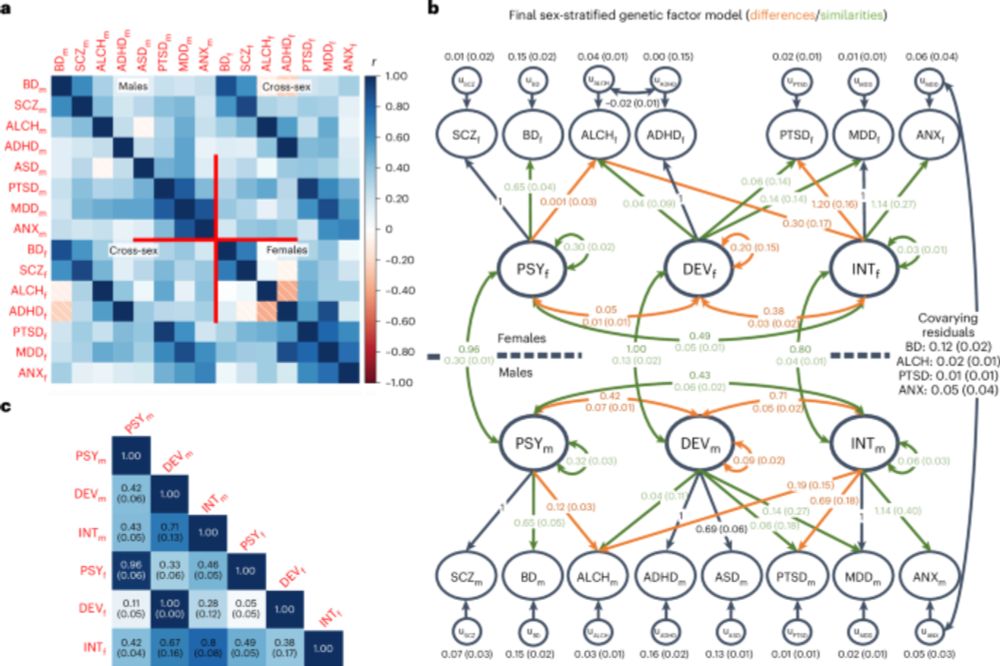
Standardized results (with standard errors) for two-factor genomic structural
equation model as estimated using GenomicSEM software.
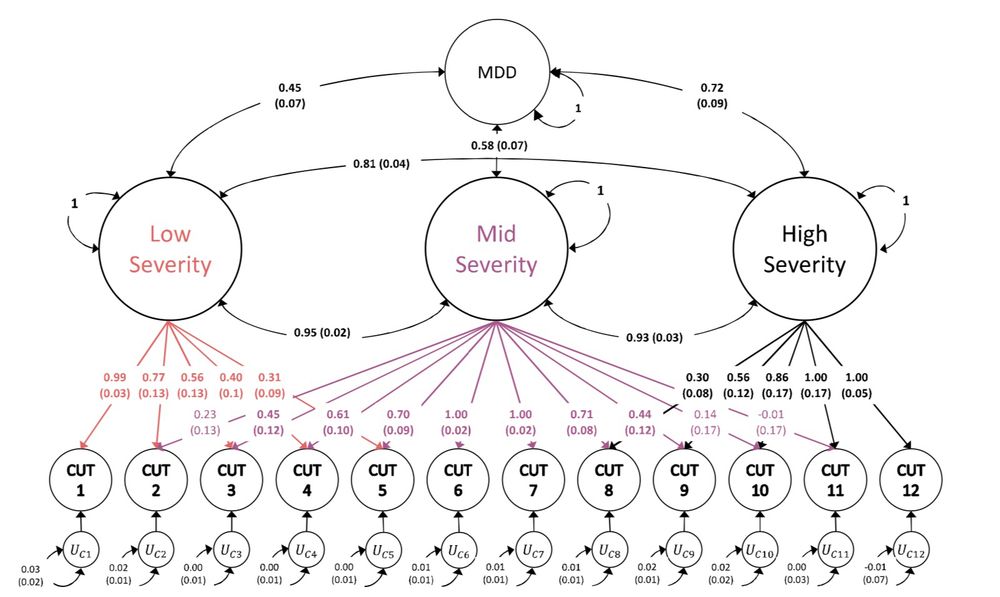
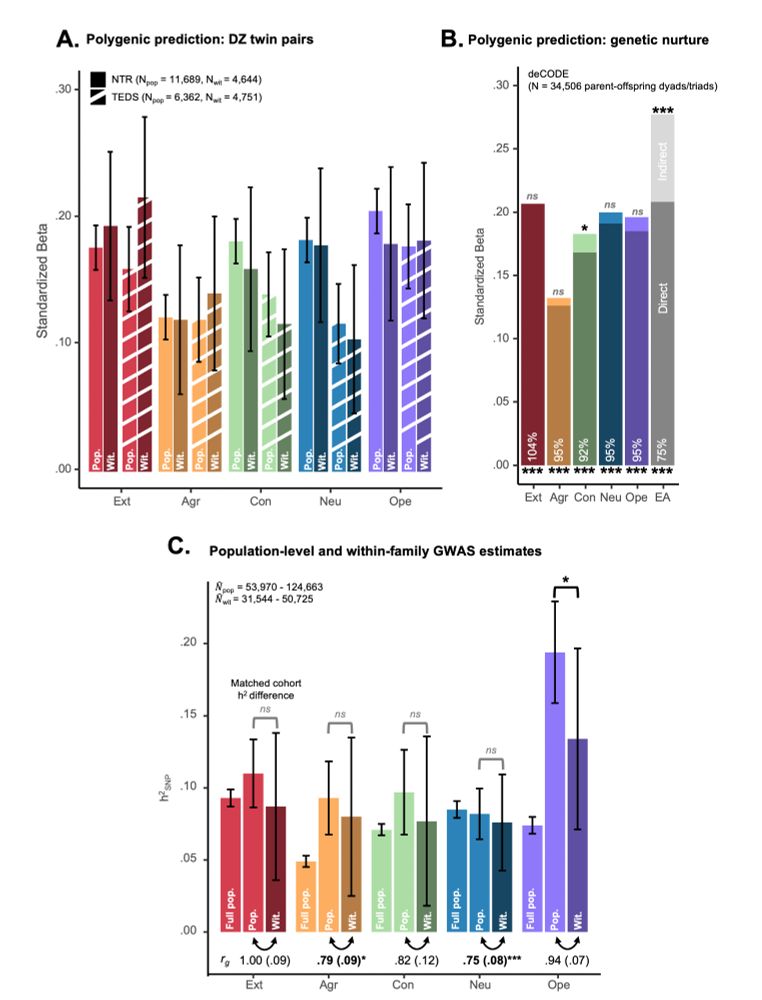
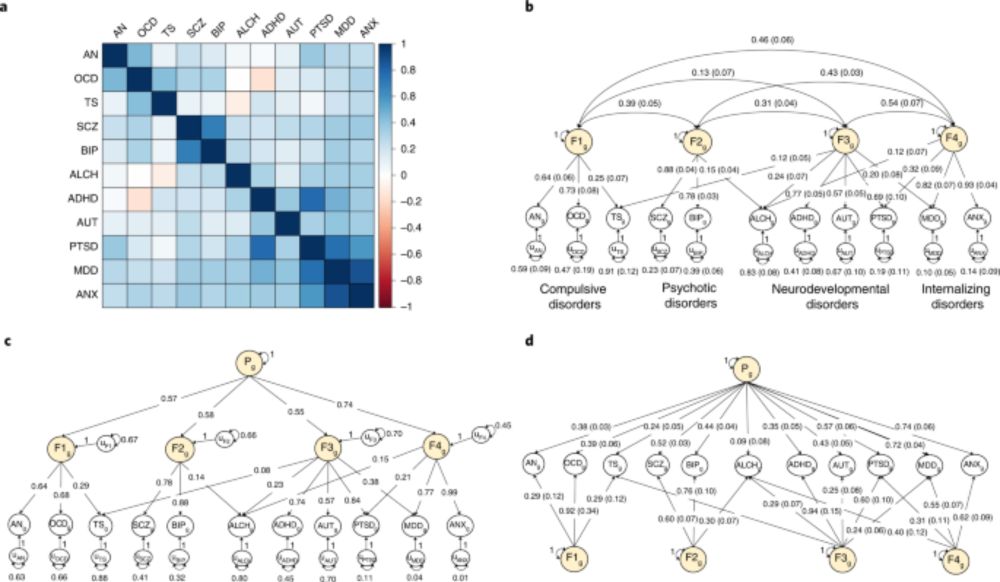
Standardized results (with standard errors) for three-severity factor genomic structural
equation model as estimated using GenomicSEM software.
Are mental disorders extreme manifestations of continuous traits or genetically distinct entities? We present Genomic Taxometric Analysis of Continuous and Case-Control data (GTACCC) for evaluating genetic continuity and differentiation of traits across the severity spectrum. Tweetorial coming soon.
05.02.2025 01:15 — 👍 34 🔁 13 💬 2 📌 2
Finally, I couldn’t end without mentioning that we were able to work in this fantastic representation of factor analysis from Buzz Hunt.
15.01.2024 17:22 — 👍 3 🔁 0 💬 2 📌 0
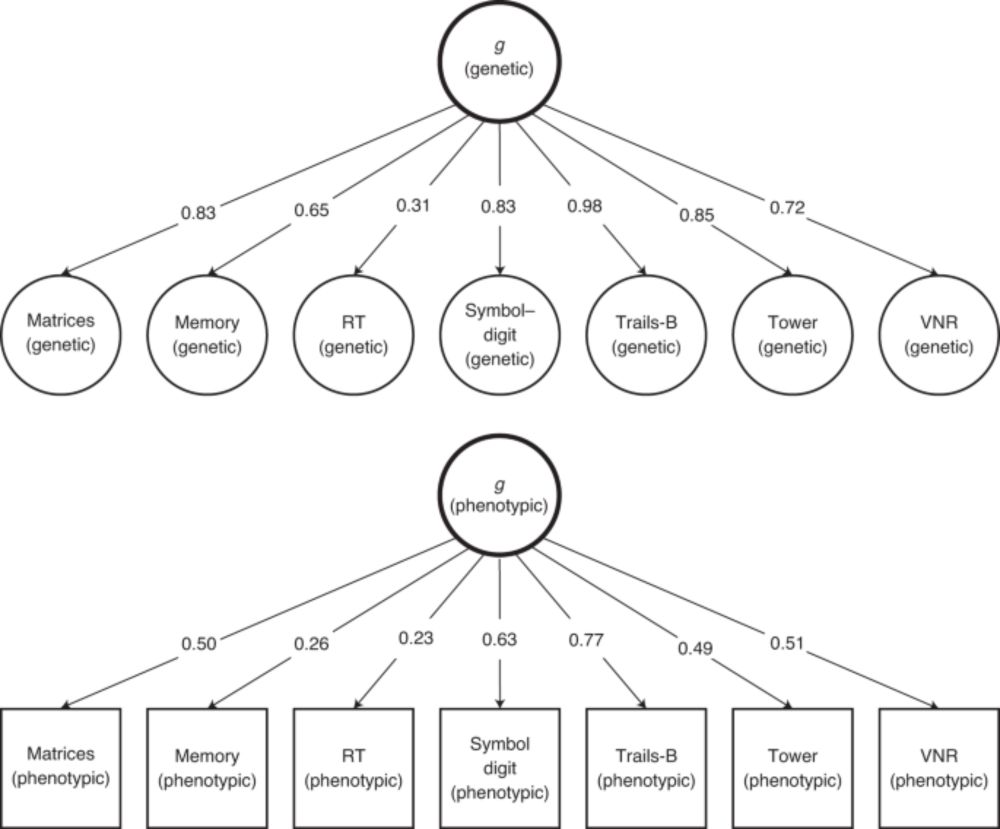
Even when the factor model is correct, we observe some variants with specific effects. Variants in the APOE region are not generally associated with all cognitive tasks. Rather, they are associated with “fluid” tasks which decline with age, but not “crystallized” tasks.
15.01.2024 17:21 — 👍 1 🔁 0 💬 1 📌 0
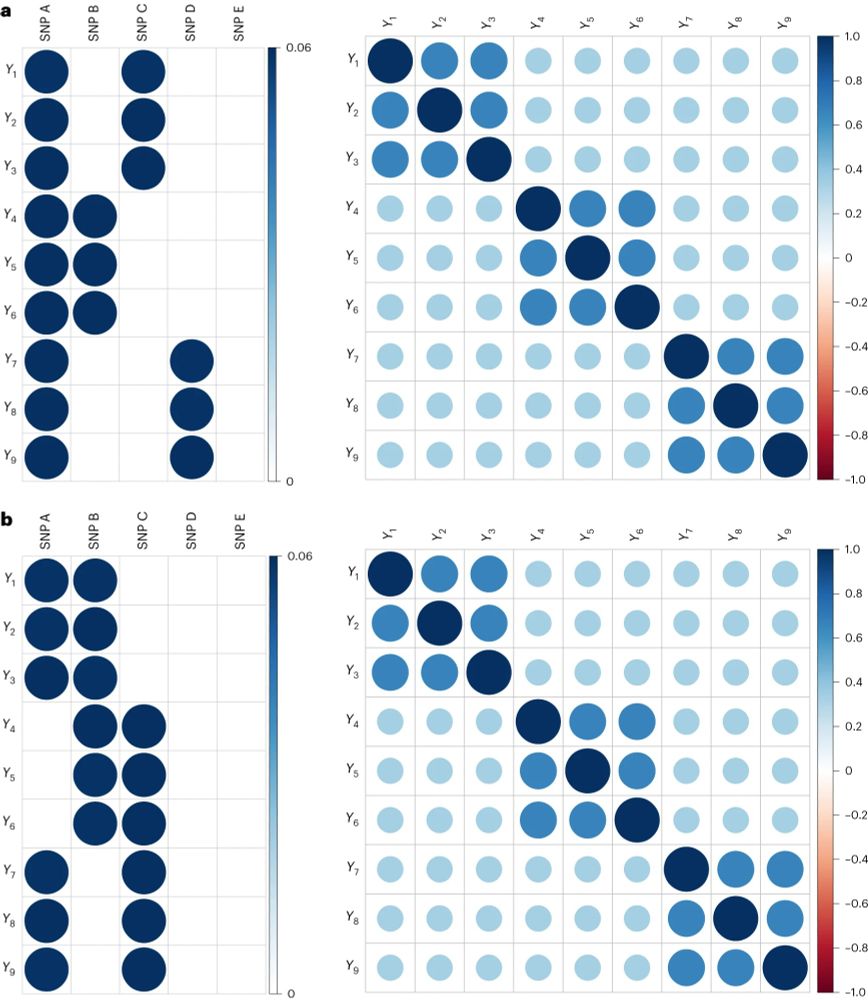
We can see this in the results of a simple simulation. Here, although the square correlations matrices (intercorrelations among phenotypes) are indistinguishable, the rectangular matrices (associations with individual genetic variants) differ starkly.
15.01.2024 17:20 — 👍 0 🔁 0 💬 1 📌 0
However, if the common factor is an illusion- a statistical artifact of aggregation- then we should observe individual genetic variants that are associated with subsets of the phenotypes, but not variants that are associated with all phenotypes.
15.01.2024 17:19 — 👍 1 🔁 0 💬 1 📌 0
If the common factor model is correct, then we should observe individual genetic variants that are associated with all of the phenotypes composing that factor (the SNP effects should be proportional to the factor loadings).
15.01.2024 17:18 — 👍 1 🔁 0 💬 1 📌 0
Genome-wide association studies (GWAS) quantify associations between genetic variants and phenotypes. We can leverage GWAS data to resolve the factor indeterminacy problem and test the validity of latent variables.
15.01.2024 17:18 — 👍 1 🔁 0 💬 1 📌 0
However, the human genome contains elementary components that, due to the shuffling process associated with sexual reproduction, come to be naturally uncorrelated.
15.01.2024 17:17 — 👍 1 🔁 0 💬 1 📌 0
Reducing cognitive tasks into smaller components was unsuccessful – these narrow components still correlated with each other, leaving open the question of what model was responsible for those correlations.
15.01.2024 17:17 — 👍 1 🔁 0 💬 1 📌 0
Researchers have suggested breaking down cognitive tasks into “elementary processes” to identify components that correlate with the phenotypes but are uncorrelated with one another. Such a set of variables could be used to test whether a general factor was the true causal model.
15.01.2024 17:17 — 👍 1 🔁 0 💬 1 📌 0
That’s factor indeterminacy: just because a set of correlations is consistent with a common factor doesn’t mean that we can rule out other types of causal models. So what do we do about factor indeterminacy?
15.01.2024 17:15 — 👍 2 🔁 1 💬 1 📌 0
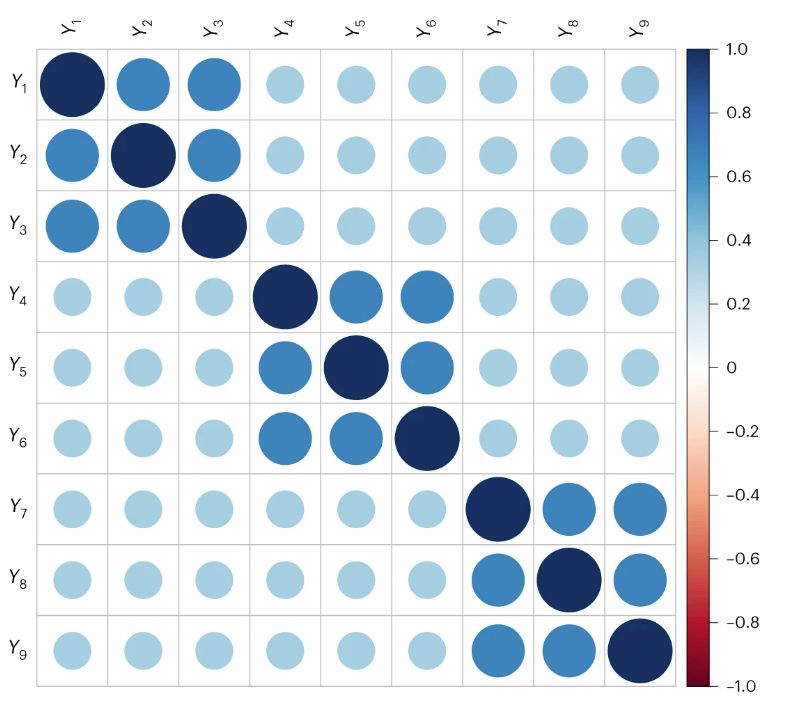
Here are correlations generated using a different model, where different combinations of 6 phenotypes share a cause, but never all 9 (so there is no common factor). It looks the same as the other matrix!
15.01.2024 17:15 — 👍 1 🔁 0 💬 1 📌 0
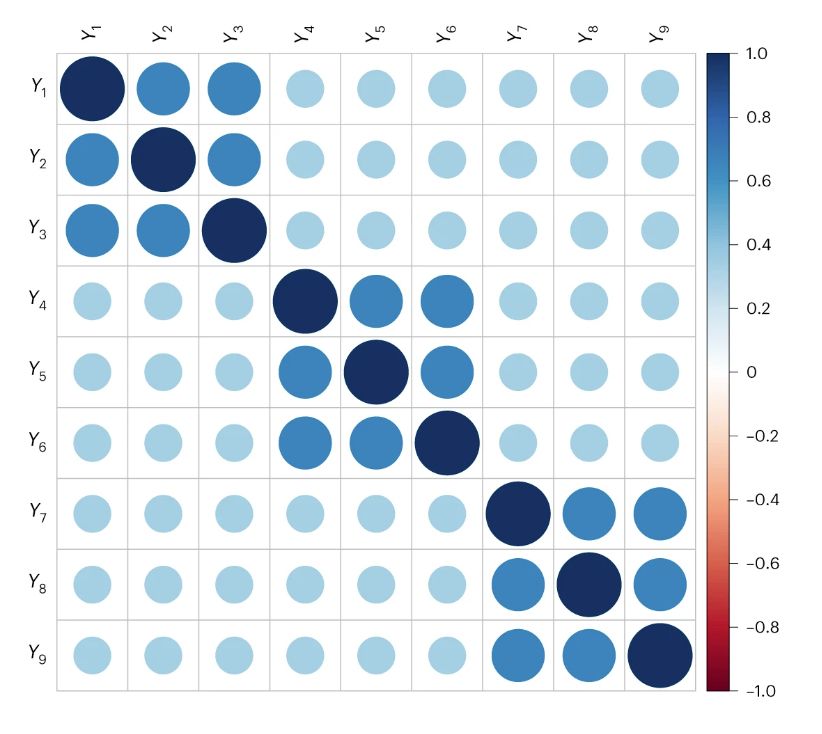
To make this a little more salient, let’s look at some correlation matrices. These matrices show the correlation among 9 phenotypes. Here’s one that was generated by a common factor model, where one shared cause influences all of the phenotypes in the correlation matrix.
15.01.2024 17:14 — 👍 1 🔁 0 💬 1 📌 0
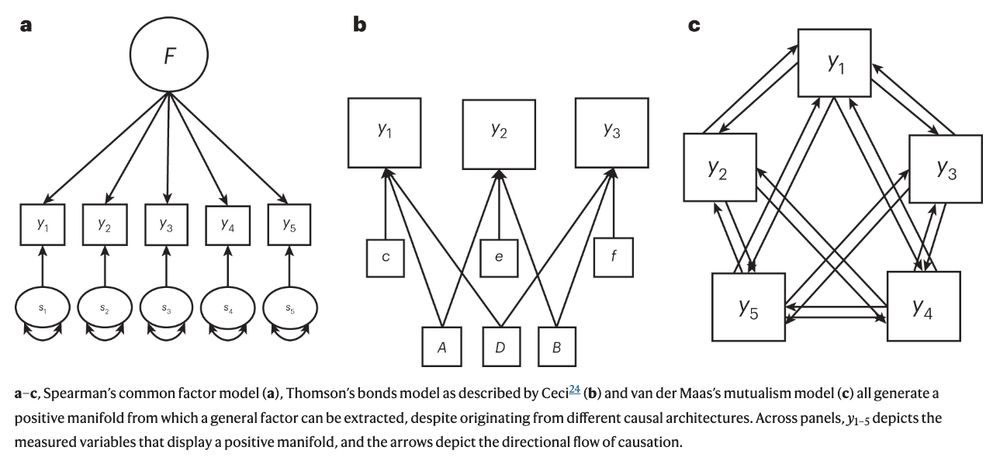
Many social scientists use latent variables to capture variables that cannot be directly observed (extraversion, intelligence). However, the correlations we use to infer latent factors can result from alternative data generating mechanisms. We refer to this problem as factor indeterminacy.
15.01.2024 17:14 — 👍 1 🔁 0 💬 1 📌 0
Beyond the factor indeterminacy problem using genome-wide association data - Nature Human Behaviour
The authors address the central criticism of latent variable models in behavioural science, which is that a wide range of causal models may account for the observed data (the factor indeterminacy prob...
🚨 New paper!
In this review, we explore factor indeterminacy and outline how we can use genetic data to test the validity of latent factors.
tinyurl.com/5n75446y
With @tedmond.bsky.social , @tuckerdrob.bsky.social , @kph3k.bsky.social , @andrewgrotzinger.bsky.social , and @michelnivard.bsky.social
15.01.2024 17:12 — 👍 25 🔁 12 💬 1 📌 2
Professional psych nerd; PhD studying friendship and mental health @goldsmithsUoL. Writes sometimes. Funnier on instagram. (he/him)
Clinical psych PhD student @ University of Virginia | Alzheimer's disease & cognitive aging | behavioral genetics & epigenetics
https://linktr.ee/roxanegay
Writer, editor, publisher, professor. Some people call me a bad feminist and by some people I mean me.
Postdoc @ IBG, Colorado. Research interests: ageing, frailty, dementia, genetic epidemiology & preventive medicine.
International society supporting education and research on complex relationships between genes, environments, and behavior: https://www.bga.org/
https://medicine.yale.edu/profile/joseph-deak/
Artist and data scientist in the Bulik and Sullivan labs (UNC SOM) investigating the underlying genetics of psychiatric disorders
PhD student at the Icahn School of Medicine at Mount Sinai. Interested in all things psychiatric genetics.
Behavioral geneticist, dad, cyclist, reluctant dog owner, formerly young, allegedly irreverent, unlike most people my age, I was raised in the Great Depression.
PhD student at @unifesp | Psychiatric Genetics | Bioinformatics | Neuroimaging | Data Viz
Clinical Science PhD Student | WashU - BRAIN Lab | I like the brain | “Reformed” Theater Kid
Is this thing on?
Physics, philosophy, complexity. @jhuartssciences.bsky.social & @sfiscience.bsky.social. Host, #MindscapePodcast. Married to @jenlucpiquant.bsky.social.
Latest books: The Biggest Ideas in the Universe.
https://preposterousuniverse.com/
Cognitive scientist, philosopher, and psychologist at Berkeley, author of The Scientist in the Crib, The Philosophical Baby and The Gardener and the Carpenter and grandmother of six.
Assistant Professor, Psychology & Data Science @ NYU | Working on brains & climate (separately) | Author of "Models of the Mind: How physics, engineering, and mathematics have shaped our understanding of the brain" http://tinyurl.com/h9dn4bw7
Paradigmatically promiscuous scientist. Modeler of cultural evolution and related topics. Professor at UC Merced and the Santa Fe Institute.
Web: https://smaldino.com/wp/
Do you really don’t know?
(I’m a philosopher and historian of biology, interested in all things evolutionary, #genetic, or #cognitive. I find most things ridiculous.)
http://www.ehudlamm.com