It is my great honor to announce that registration for the 7th ESCP Single Cell Proteomics Conference is now open: lnkd.in/e4iyiQjf
With around 250 participants, it is one of the largest SCP conferences worldwide. We are also proud to announce that there is no participation fee for our conference.
28.01.2026 16:23 — 👍 18 🔁 10 💬 2 📌 1

Single cell proteomic analysis defines discrete neutrophil functional states in human glioblastoma
Neutrophils are vital innate immune cells shown to infiltrate glioblastomas, however we currently lack the molecular understanding of their functional states within the tumour niche. Neutrophils are k...
Happy to share our latest preprint doing low cell number (mini-bulk) and single cell #proteomics on tumour associated neutrophils from human glioblastoma where we find multiple functional states that would be invisible to scRNAseq, some showing pro-tumoural states with potential therapeutic value
27.07.2025 12:28 — 👍 64 🔁 18 💬 3 📌 3
If it only live for 12-24 hours, how we can call it senescence?
03.05.2025 07:00 — 👍 1 🔁 0 💬 1 📌 0
Learn most of proteomics knowledge from MaxQuant training videos on YouTube
21.03.2025 03:01 — 👍 1 🔁 0 💬 0 📌 0

Exciting to share a new low-input proteomics technology for unbiased profiling cell surfaceome. The nanoMAPS miniaturizes bead-based AP-MS inside a single droplet, a key step in extending sensitive proteomics from global to functional measurement. www.biorxiv.org/content/10.1...
16.03.2025 23:04 — 👍 30 🔁 5 💬 1 📌 2
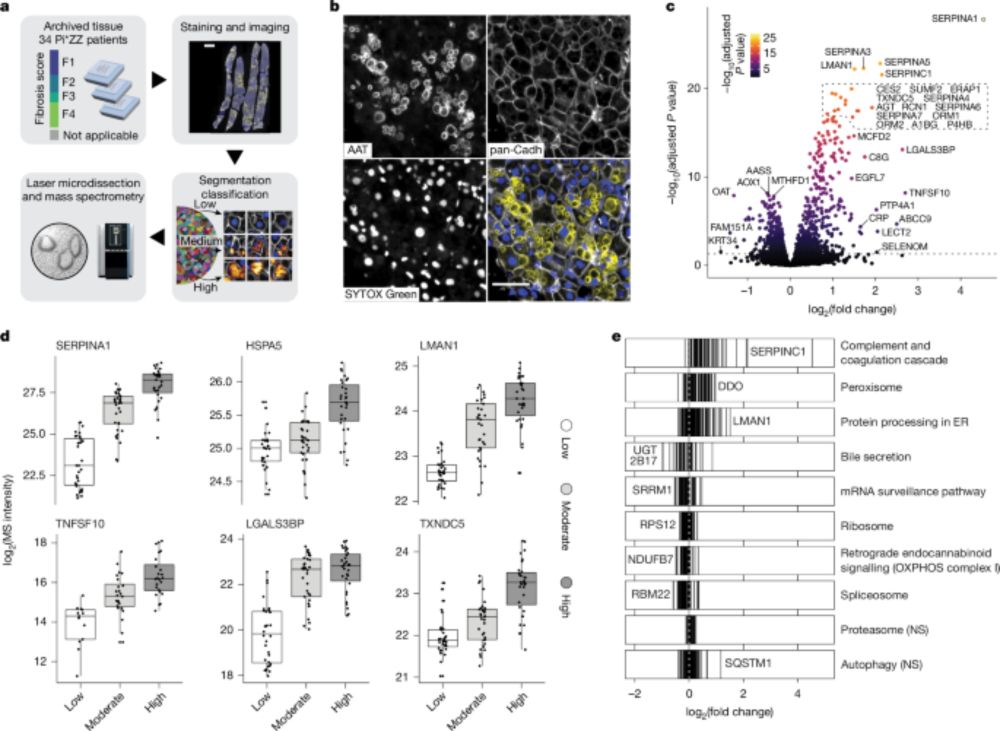
Very important study to allow the light cell fixation for single cell proteomics!!! Results are very promising!
04.02.2025 18:21 — 👍 7 🔁 1 💬 0 📌 0
Welcome!
17.01.2025 07:56 — 👍 1 🔁 0 💬 0 📌 0
Perfect couple of DESI with microPOTS spatial proteomics! Glad to see microPOTS and DDM protocols received more uses
22.12.2024 23:46 — 👍 4 🔁 0 💬 0 📌 0
DDM may not the best to make bubbles, but definitely the best to get your peptide off from the tubes
20.12.2024 08:28 — 👍 3 🔁 0 💬 0 📌 0
We Are the Analyzers - A Proteomics Anthem by US HUPO.
We Are the Analyzers - Proteomics Anthem
We’re thrilled to present "We Are the Analyzers" – an anthem for the proteomics community! A huge shoutout to everyone who brought this incredible song to life. Look what proteomics has already shown us 🎶🧪youtu.be/wzmJDNmsWK8 #Proteomics #USHUPO20YRS
18.12.2024 19:00 — 👍 22 🔁 12 💬 1 📌 4
The main idea behind this is to use transcript reference mapping to assign the cell types, and then do proteomics DE analysis to avoid the double dipping. So external single cell databases also take into considerations
06.12.2024 07:26 — 👍 7 🔁 0 💬 1 📌 0
After long-time review, #nanoSPLITS is finally published. I am deeply grateful to my colleagues who made this work possible, especially @cajunscience.bsky.social. I am also grateful to the support of our single-cell proteomics community during the peer review on Nature Communication!
06.12.2024 00:41 — 👍 35 🔁 7 💬 1 📌 0
thanks for post it!
06.12.2024 00:36 — 👍 1 🔁 0 💬 0 📌 0
I have more questions on Figure 4. Seems IonQuant and Maxquant can not give correct estimates of spike-in protein abundance. I am not sure if it is a normalization issue or FDR control issue.
04.12.2024 05:51 — 👍 6 🔁 0 💬 0 📌 0
It is a great/beautiful study to push the low input proteomics to clinical space. I do agree the title is misleading, it should name as “single cell type proteomics” or “cell type specific proteomics”
03.12.2024 19:49 — 👍 2 🔁 0 💬 0 📌 0
Congratulations @proteomicsnews.bsky.social, can not wait to see more exciting single cell studies from your lab
28.11.2024 02:52 — 👍 1 🔁 0 💬 0 📌 0
6th ESCP 2025 – APMA
It gives me great pleasure to announce our next European Single Cell Proteomics Conference www.apma.at/6thescp/
We would be delighted to receive many abstracts from students so that they have the opportunity to give their first presentation.
On behalf of: Erwin, Fabian, Manuel, Fabian and Karl
21.11.2024 07:14 — 👍 27 🔁 14 💬 0 📌 3
As I moved my focus to more scientific questions, I prefer to have less identification, better quantification, and more trustworthy data. It is super expensive to validate each proteins for downstream study
18.11.2024 04:26 — 👍 3 🔁 0 💬 1 📌 0
Cool
17.11.2024 03:09 — 👍 1 🔁 0 💬 0 📌 0
I will try to get him here
17.11.2024 02:38 — 👍 3 🔁 0 💬 1 📌 0
Such a tool is in great need. Will dig into this paper! Thanks for sharing
16.11.2024 22:03 — 👍 1 🔁 0 💬 0 📌 0
The Mann Lab is a pioneer in mass spectrometry-based proteomics. Posts represent personal views from lab members and Matthias Mann
Absea Biotechnology | Empowering protein science
14,000+ recombinant proteins & 11,000+ monoclonal antibodies covering 70% of the human proteome.
Germany-USA-China
https://www.absea.bio/index.html
Postdoc at UCSF Wells lab, PhD 22’ Harvard Woo lab, BS 17’ Peking University Peng Chen lab
Proteomics, Mass Spectrometry. Group leader at Institute of Systems Medicine, Chinese Academy of Medical Sciences.
VP & Head of Induced Proximity Platform, @Amgen | Scientist | Father
I am a scientist. #imaging #massspec #biofilms 🧫 #ovariancancer #microbial communities 🧀 Prof of Chemistry and Biochemistry @ucsantacruz.bsky.social she/her
Proteomics, precision health, mass spectrometry, biomarkers, quantitative, changing medicine
Tenure-track Assistant Prof at University of Copenhagen | Mass spectrometrist and hobbyist elephant flyer | East Van boy at heart | Views are my own. 🇨🇦 in 🇩🇰
Assistant professor of chemistry at U of Utah
www.nagylab.com
Group leader at Leibniz-Forschungsinstitut für Molekulare Pharmakologie
W2-S Professor at Charité – Universitätsmedizin Berlin
Science: Group Leader at WEHI, Immunology, Haematopoiesis, Dendritic cells, Clonal lineage tracing, Barcoding
Media: Ask the Doctor, Catalyst, The Jab Gab, Searching for Superhuman, The Great Acceleration
Senior Scientist at Princess Margaret Cancer Centre, Toronto, Canada 🇨🇦
Clinical proteomics, liquid biopsies and cell surface markers
Professor of Cell & Regenerative Biology and Chemistry at UW-Madison, Director of Human Proteomics Program🌱 | Passionate about Top-Down Proteomics, Cardiac Systems Biology, and Precision Medicine 🔬 | Optimist who loves gardening 🌸 Views are my own.
Science integrity consultant and crowdfunded volunteer, PhD.
Ex-Stanford University. Maddox Prize/Einstein F Award winner
NL/USA/SFO.
#ImageForensics
@MicrobiomDigest on X.
Blog: ScienceIntegrityDigest.com
Support me: https://www.patreon.com/elisabethbik
PhD Student at the Cell Diversity Lab - DTU interested in mass spectrometry, single-cell proteomics and outdoor adventures
Chemistry Professor at Iowa State University. Mass spectrometry
Professor, Cedars-Sinai Med. Center. Focus: Cell Fate Mechanisms, Novel Genetic Toolsets, Tumor Models, and Neural Protection. Opinions are my own.
Mass spectrometrist at heart currently with the Broad Institute
Laboratory for Immunochemical Circuits at the La Jolla Institute for Immunology. Dept. of Pharmacology and Moore's Cancer Center, UCSD. Global Autoimmune Institute Assistant Professor. samyerslab.org