GitHub - HiDiHlabs/sainsc: Segmentation-free Analysis of In Situ Capture data
Segmentation-free Analysis of In Situ Capture data - HiDiHlabs/sainsc
Sainsc is a new tool for efficient whole organism spatial transcriptomics data analysis at the nanometre scale. It works with Stereo-seq, VisiumHD, Xenium and Nova-ST. Looking forward to see how it works with upcoming platforms (e.g. Illumina).
github.com/HiDiHlabs/sa...
04.07.2025 05:47 — 👍 1 🔁 0 💬 0 📌 0
And our tireless worker bees who dedicated every Wednesday evening for 18 months for this project: Jieran Sun, Kirti Biharie, Peiying Cai, Niklas Müller-Bötticher, Paul Kiessling, Meghan A. Turner, Søren H. Dam, Florian Heyl. 🙏
03.07.2025 15:47 — 👍 0 🔁 0 💬 0 📌 0
I forgot the most important bit! This would not have been possible without the wonderful and amazing SpaceHack community. Special shout outs to Brain Long, Ahmed Mahfouz and @markrobinsonca.bsky.social for their work, ideas and resources...
03.07.2025 15:44 — 👍 1 🔁 0 💬 1 📌 0
GitHub - SpatialHackathon/SACCELERATOR
Contribute to SpatialHackathon/SACCELERATOR development by creating an account on GitHub.
We wrap up our analysis framework into SACCELERATOR, github.com/SpatialHacka..., and present two case studies on brain and colon cancer datasets to remind people that having domain expertise and performing interactive analysis is more important than picking the "best" tool.
02.07.2025 12:14 — 👍 1 🔁 0 💬 1 📌 0
General findings:
- benchmarking studies can be inconsistent
- methods do not scale to new datasets
- "ground truths" do not consider granularity of biological features
- looking at the differences between methods (entropy) is informative
- method performance doesn't match subjective importance
02.07.2025 12:11 — 👍 2 🔁 0 💬 1 📌 0

2D, or not 2D? Investigating Vertical Signal Integrity of Tissue Slices
Imaging-based spatially resolved transcriptomics can localise transcripts within cells in 3D. Cell segmentation precedes assignment of transcripts to cells and annotation of cell function. However, ce...
Have you been bothered that practically every #SpatialTranscriptomic analysis tool assumes data to be 2D? Well, you were right - overlapping cells are a really big problem. Check out our report on bioRxiv: 2D, or not 2D? Investigating Vertical Signal Integrity of Tissue Slices. t.ly/CQX8K
08.02.2025 09:43 — 👍 7 🔁 3 💬 1 📌 0

GitHub - HiDiHlabs/multiSPAETI: Implementation of MULTISPATI-PCA in Python
Implementation of MULTISPATI-PCA in Python. Contribute to HiDiHlabs/multiSPAETI development by creating an account on GitHub.
We also sneak in a python implementation for the spatially aware MULTISPATI-PCA. We think this should pretty much replace "non-spatial" PCA for all spatial modalities in most cases.
GitHub: github.com/HiDiHlabs/mu...
RtD: multispaeti.readthedocs.io/1.0.0/genera...
08.02.2025 09:40 — 👍 1 🔁 0 💬 0 📌 0
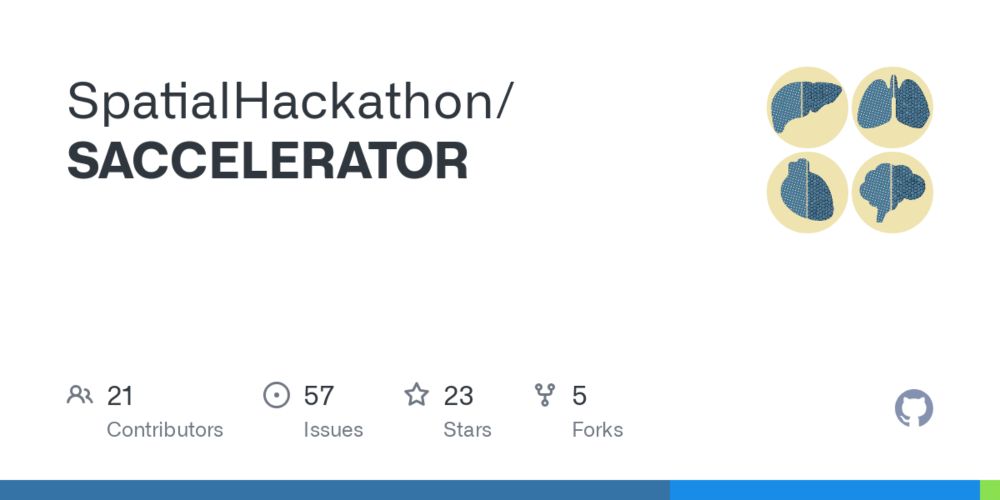
SpatialLeiden: spatially aware Leiden clustering - Genome Biology
Clustering can identify the natural structure that is inherent to measured data. For single-cell omics, clustering finds cells with similar molecular phenotype after which cell types are annotated. Le...
SpatialLeiden is out in #GenomeBiology: t.ly/WQppF. It models both gene expression and spatial information for clustering. This allows you to perform e.g. spatially aware cell typing or domain identification. It's fast, scalable and intuitive for single-cell researchers. Try it out!
08.02.2025 09:33 — 👍 3 🔁 1 💬 2 📌 0
The HISTOMAP projects aims to use AI and spatial transcriptomics data to accelerate biomarker detection for bladder cancer. We will work closely with the clinic, pathology, and regulatory experts to investigate how best to get these models into clinical routine.
08.01.2025 10:15 — 👍 0 🔁 0 💬 0 📌 0
I tried Sainsc for our MERSCOPE data and Naveed, it works wonderfully! Importantly, the documentation is excellent. Thank you so much for the clear tutorial and explanations. This is truly a life saver for my analysis at the moment. Props to the team, this is awesome work!
05.12.2024 09:42 — 👍 1 🔁 1 💬 0 📌 0
Everyone knows by now that 42 is the answer to the ultimate question of life, the universe, and everything. But as I turn 42 today, I’ve realised something profound: it’s not the answer that matters - it’s the prompt!
19.12.2024 19:26 — 👍 3 🔁 0 💬 0 📌 0
Am I really the first to repost Mats Nilsson? What an honour!
17.12.2024 09:24 — 👍 1 🔁 0 💬 0 📌 0

a cartoon of a man looking out a window at the stars
ALT: a cartoon of a man looking out a window at the stars
Hello world. Our lab has a new home here on Bluesky! We will post about spatial omics, science, nice papers and lab activities. The account will be collectively managed by lab members. Happy spatial-omics everyone!
12.12.2024 16:24 — 👍 5 🔁 1 💬 1 📌 0

The calm before the storm... #SpaceHack starts tomorrow! Looking forward to 3 intense days of addressing unsolved data analysis problems for #SpatialTranscriptomics.
More here: spatialhackathon.github.io
08.12.2024 17:27 — 👍 11 🔁 4 💬 0 📌 0
SpatialLeiden adds spatial support to the Leiden clustering algrithm. It has 3 parameters. Two we know from scRNAseq analysis (resolution and num of neighbors), and the other is the weight of the spatial info. We thin it is the natural choice for spatial clustering for the single-cell community!
03.12.2024 20:03 — 👍 0 🔁 0 💬 0 📌 0
Home
Github Pages for SpatialHackathon
... we will also implement multi-omics and multi-sample support at #SpaceHack spatialhackathon.github.io
03.12.2024 19:56 — 👍 0 🔁 0 💬 0 📌 0
@gestaltsp.bsky.social welcome to BlueSky!
@jasmineplummer.bsky.social @ioavlachos.bsky.social
29.11.2024 09:59 — 👍 6 🔁 2 💬 1 📌 0
VisiumHD: From 16 µm to 2 µm resolution — sainsc 0.2.2.dev5+g0c33c57 documentation
Have you performed a #VisiumHD run, and frustrated that you had to analyse data in 8/16 um bins?
We just updated #Sainsc to create maps of cell types at 2 um resolution for #VisiumHD: sainsc.readthedocs.io/tutorials/tu....
Check it out and let us know - happy to add features/fixes.
29.11.2024 10:32 — 👍 5 🔁 1 💬 0 📌 0
#Sainsc is a cell-segmentation-free tool to create cell-type maps.
Why segmentation-free? Cell segmentation is hard, and if not done well it can ruin expression profiles, even miss cells.
Sainsc is fast! Get first insights in <5 min for #Xenium, #MERSCOPE, and <1 hr for #STomics, #Stereo-seq.
29.11.2024 10:29 — 👍 1 🔁 0 💬 0 📌 0

Whole-brain spatial transcriptional analysis at cellular resolution
Incredible study - 3D single cell imaging of the entire mouse brain - also on the cover of this week's #Science
🧪🔬🤯
www.science.org/doi/10.1126/...
22.11.2024 16:16 — 👍 65 🔁 18 💬 0 📌 2
Id love to be added :)
24.11.2024 22:15 — 👍 1 🔁 0 💬 0 📌 0
🇨🇦 boiling in the Arizona desert and not riding my bikes enough. Cancer researcher focused on multiple myeloma and clinical genomics at TGen, part of City of Hope. Creator of MyelomaSky and Sequencing Tech feeds. Director, Collaborative Sequencing Center
Senior data scientist in computational structural biology, protein dynamics and cryoem | Marie Sklododowska-Curie fellow w Jose Maria Carazo and Carlos Oscar Sorzano | postdoc w Ivet Bahar
Associate Professor
DFCI & HMS
Assistant Professor at UT Southwestern Medical Center | Full-time reproductive biologist | Part-time genomics technology developer | Lab website: https://haiqichenlab.github.io/
Head of the Centre for Artificial Intelligence, DS&AI, Biopharma R&D, AstraZeneca Views are my own
Associate Director in Bioinformatics and Data Science at Remix Therapeutics.
Biotech. Lives in Waltham, Massachusetts.
🇸🇪
Tenure-Track-Professor for Multifactorial Data Analysis and Machine Learning in the Life Sciences @UniHeidelberg
https://velten-group.org/
Spatial-omics nerd in NYC looking for new ways to connect scientists with the tools they need.
#Trekker
PhD student in the Saez-Rodriguez group (@saezlab.bsky.social) @ebi.embl.org. Member of Gonville & Caius College (@caiuscollege.bsky.social).
Computational biology | Omics | Microbiome | IBD & CRC | Open science
Fighting blood cancer🩸with a pipet + Senior Postdoc 🧪🧬 + Berlin + ❤️ cœur perdu à Paris 🇫🇷 + Feminist + mom of 👦🧒 & 🐰🐰+ FirstGen + she/her + not nice + Politics and SciCom mostly in German + Tax the rich
I have kids in this world.
I‘m allowed to be mad.
PhD student in Statistical Bioinformatics at University of Zurich and SIB
PhD student, Neuroscience, CSIR Fellow, Investigating Alzheimer's disease crosstalk with metabolic syndrome | Using Bioinformatics, Machine Learning and AI in research.
#Braindisease #Drugdiscovery #Academia
Innovative platform optimizing biological sample management in research, promoting animal welfare and reduction. 🌍🔬
🔸 https://arukon.com/
✉️: arukoninfo@gmail.com
#AnimalResearch
Scientist studying epithelial biology, stem cells and cancer at UCL (@epicentr.bsky.social) 🫁
Statistical geneticist. Professor of Human Genetics and Biostatistics at the University of Pittsburgh. Assiduously meticulous.
Interpretable ML | Computational and Systems Biology | Postdoc at Cancer Early Detection Advanced Research (CEDAR) Center @ohsuknight.bsky.social