Fascinating link between gene biosynthesis and cuproptosis
21.09.2025 02:15 — 👍 0 🔁 0 💬 0 📌 0
Spearheaded by
@jeffreychsiao.bsky.social and Doug Warui 🙌 . Huge thanks to Jason Kwon for the computational analysis , Tamra, @maggiedreishpoon.bsky.social , Nolan Bick and
Squire Booker for making this possible.
@bidmc-cancercenter.bsky.social @broadinstitute.org
13.09.2025 14:07 — 👍 1 🔁 0 💬 0 📌 0
Key take-home points:
1.The acidic α-helix 3 is a crucial regulatory interface on FDX1
2. The roles of FDX1 in cuproptosis and lipoylation couldn't be uncoupled, suggesting they are a single, structurally linked function
3.DLD is a newly discovered upstream reductase for FDX1
13.09.2025 14:07 — 👍 0 🔁 0 💬 1 📌 0

Indeed, DLD bound FDX1 more strongly than FDXR in cells, and this interaction was lost with the D136R/D139R mutants. DLD could also replace FDXR in an FDX1-dependent lipoylation assay in vitro .
13.09.2025 14:07 — 👍 0 🔁 0 💬 1 📌 0
This hinted that FDX1 might need or have another electron donor in cells (which is not FDXR). Based on structural homology, we tested if DLD (dihydrolipoamide dehydrogenase), the E3 subunit of lipoylated complexes, could also serve as an FDX1 electron donor.
13.09.2025 14:07 — 👍 0 🔁 0 💬 1 📌 0

But there was a twist. Cells with D136R or D139R mutants were also completely deficient in lipoylation. Yet in vitro these mutants stayed fully functional — supporting LIAS-mediated lipoylation and also binding FDXR/LIAS with similar affinities as WT.
13.09.2025 14:07 — 👍 0 🔁 0 💬 1 📌 0

This analysis highlighted two residues — D136 and D139 on FDX1’s α-helix 3.
Mutating them to alanine left cuproptosis intact, but flipping their charge (Asp → Arg) made cells resistant to elesclomol–Cu–induced death.
13.09.2025 14:07 — 👍 0 🔁 0 💬 1 📌 0

This let us filter out mutations predicted to disrupt overall folding. We focused on variants that blocked FDX1-driven cuproptosis while predicted to remain structurally intact.
13.09.2025 14:07 — 👍 0 🔁 0 💬 1 📌 0

We paired the DMS data with an in silico FoldX analysis, which predicts the mean free-energy change (ΔΔG) for each residue in FDX1.
13.09.2025 14:07 — 👍 0 🔁 0 💬 1 📌 0

To find out, we performed deep mutational scanning (DMS), mutating every amino acid in FDX1 and testing its ability to promote cuproptosis in two cell line models.
13.09.2025 14:07 — 👍 0 🔁 0 💬 1 📌 0
So FDX1 has two roles: 1. Directly reducing Cu(II) bound to elesclomol, freeing Cu(I) and 2. Promoting protein lipoylation, the copper target in cuproptosis.
We set out to explore if these functions could be structurally uncoupled.
13.09.2025 14:07 — 👍 0 🔁 0 💬 1 📌 0

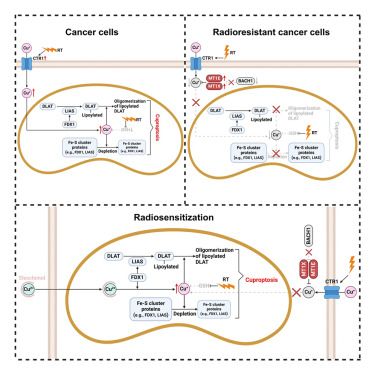
Copper induces cell death by targeting lipoylated TCA cycle proteins
Lipoylation determines sensitivity to copper-induced cell death.
We previously showed that #FDX1 is essential for #cuproptosis , a copper-dependent cell death pathway where Cu targets lipoylated enzymes & Fe–S cluster proteins. www.science.org/doi/10.1126/...
13.09.2025 14:07 — 👍 0 🔁 0 💬 1 📌 0
New preprint out!
Deep Mutational Scanning of FDX1 Identifies Key Structural Determinants of Lipoylation and Cuproptosis
A quick twitorial 🧵
www.biorxiv.org/content/10.1...
13.09.2025 14:07 — 👍 1 🔁 1 💬 1 📌 0
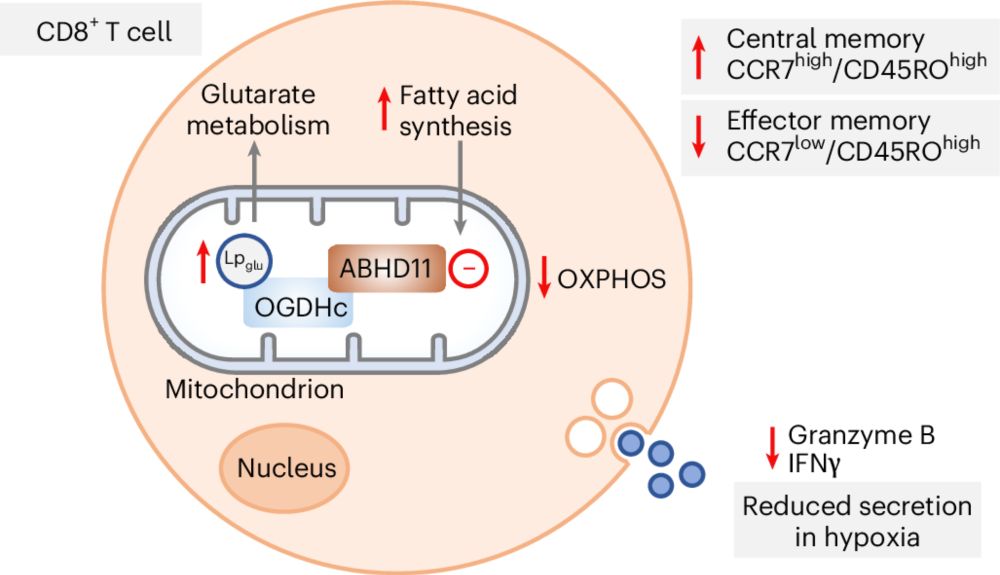
Lipoyl deglutarylation by ABHD11 regulates mitochondrial and T cell metabolism
Nature Chemical Biology - Glutarate rewires cell metabolism through conjugation to proteins or fatty acids, but how glutarylation is regulated is unclear. ABHD11 was identified as a...
Delighted to share our latest work on ABHD11 as a novel deglutarylating enzyme, and its role in T cell metabolism @natchembio.nature.com rdcu.be/ewiYZ. Many congratulations to co-first authors Guine Grice, Eleanor Minogue and @hudsonwilco.bsky.social! Great team effort with Randall Johnson's lab.
15.07.2025 16:12 — 👍 20 🔁 5 💬 1 📌 0

Image credit: Deqiang Kong, Yue Wang and Tianxing Yan. Cover design: Bethany Vukomanovic
❤️Our May issue is out❤️Read our three articles focused on atherosclerosis, describing TLO-like structures within plaques, FDX1’s protective role, and how gut microbiota-derived bile acids suppress platelet activation, and more!
www.nature.com/natcardiovas...
19.05.2025 13:26 — 👍 4 🔁 2 💬 1 📌 0
Just had exactly the same experience - went to watch wicked with my girls -with the exact opposite outcome, so boring. lucky they had beer in the theater 😂 .
25.12.2024 04:20 — 👍 2 🔁 0 💬 0 📌 0

chalk + chalkboard with diagrams

lab group silly picture
Wow - bluesky! 📈 🦠
Please help me amplify a special faculty search. We're looking for someone in "Microbial Systems Biology" to join me and @kreynoldslab.bsky.social at UTSW Sys Bio / Bioinformatics.
There is an empty lab space right next to us waiting for you!
apply.interfolio.com/154339
1/4
22.11.2024 17:49 — 👍 93 🔁 104 💬 3 📌 0
UK orchid conservation charity | The Brickell Award 2024 | RHS Gold medal winners 🏆| #🌿Six National Collections of orchids | 📍West Sussex, UK
Cancer Cell provides a high-profile forum to promote major advances in cancer research and oncology. We're thinking about cancer from a holistic perspective and finding ways to bridge the gap between the bench and the clinic.
https://www.cell.com/cancer-c
A laboratory of Systems Pharmacology at UMass Medical School. We study cancer therapies, and aim to understand how they activate cell death and how to make this work better.
https://www.umassmed.edu/Lee-Lab
Head of the Budin lab at UCSD (www.budinlab.com). Musings on thin layers of grease in our cells (and other topics). Lipids, cell membranes, biophysics, chem bio, evolution. 🏳️🌈
Group Leader @ Janelia Research Campus, protein engineer, 🏳️🌈 she/they
Sharing speeds science! Addgene helps scientists around the world find and share research materials - plasmids, viral preps, antibodies, data, and more.
Semper vestigans | All things mitochondria
https://biochem.unl.edu/oleh-khalimonchuk-lab/
@Stanford, we study mitochondrial dynamics, transport, behaviors, signal transductions, and contacts in brain cells.
http://xinnanwanglab.stanford.edu
Swiss-Belgian biologist, assistant professor @UNIL with interest in mitochondria, energy metabolism, innate immunity and systems biology.
Non-profit publisher of @embojournal.org, @emboreports.org, @embomolmed.org, @molsystbiol.org and @lsajournal.org. #OpenScience #Policy
A BlueSky feed for the blog http://covalentmodifiers.blogspot.com. Discusses the covalent modification of proteins, particularly with regards to inhibitors.
A network of around 800 biochemical scientists from academia and industry founded in 1981, including around 45% students and young members. @gdch.bsky.social https://en.gdch.de/network-structures/gdch-structures/biochemistry.html
Talus Bio is a venture-backed early-stage biotechnology startup company working on drug development for gene regulators.
My lab studies the interplay between cancer metabolism and cell cycle dysregulation. Professor at The Wistar Institute
airdlab.com
Improving patient care through cutting-edge research 💊
Fusing science, medicine, & engineering to revolutionize drug discovery and treatment since 2014 🔬
labsyspharm.org | linkedin.com/company/laboratory-of-systems-pharmacology
Cell Death and Immunity Laboratory. We are interested in the mechanisms of #Apoptosis, #Necroptosis and #Inflammation in #Cancer. Posts by @ttenev.bsky.social
Howard Hughes Medical Institute (HHMI) believes in the power of individuals to advance science through research and science education, making discoveries that benefit humanity.
A leading center for expert, compassionate cancer care and advanced research.
Appts: 877-442-3324 | http://dana-farber.org
The Sarosiek Lab @HarvardChanSPH studies the regulation of #apoptosis in human development and diseases, especially cancer.