Happy to see this work published! Thanks to everyone who contributed 🙏
13.10.2025 08:11 — 👍 1 🔁 1 💬 0 📌 0
Dusa’s study is on the cover of this month’s #CellGenomics! Check it out if you haven’t had the chance yet!
www.cell.com/issue/S2666-...
10.10.2025 09:32 — 👍 5 🔁 1 💬 0 📌 0

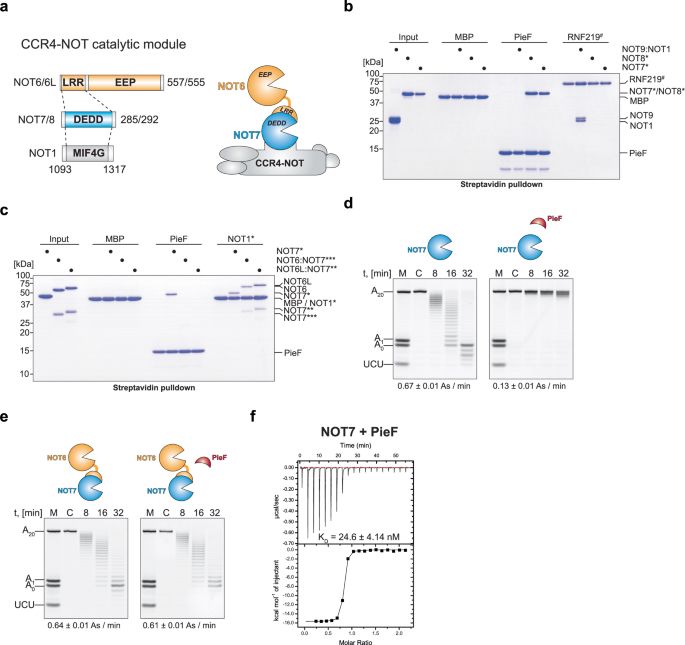
🛑Guille's @guillepalou.bsky.social no-nonsense 😉 paper is out in @springernature.com Genome Biology!
We report variable activity of the NMD pathway (mRNA QC) across tissues & individuals:
🧠less active in the nervous system & in reproductive tissues
🩸more active in digestive tract, muscle, blood
29.09.2025 16:54 — 👍 5 🔁 2 💬 1 📌 0
🚀 Our new paper is out @natmethods.nature.com!
Kuffer & Marzilli engineered conditionally stable MS2 & PP7 coat proteins (dMCP & dPCP) that degrade unless bound to RNA, enabling ultra–low-background, single-mRNA imaging in live cells.
🔗 www.nature.com/articles/s41...
🧬 www.addgene.org/John_Ngo/
22.09.2025 18:27 — 👍 223 🔁 97 💬 6 📌 9
wrong bioRxiv link here, sorry.
the correct link to the collaboration with Torben and Clemens is:
www.biorxiv.org/content/10.1...
17.09.2025 16:55 — 👍 7 🔁 2 💬 0 📌 0
NMDRHT v1.2
The team of Prof. Dr Niels Gehring @volkerboehm.bsky.social records a comprehensive database that records systematically which genes and gene variants are directly affected by "Nonsense-mediated mRNA Decay" which opens up new avenues for RNA and genome research.
The database ➡️ nmdrht.uni-koeln.de
12.09.2025 12:50 — 👍 6 🔁 4 💬 1 📌 0
🥳 Congratulations from #CRC1678 to @volkerboehm.bsky.social and the Gehring lab for their publication in @cp-molcell.bsky.social on how “Rapid UPF1 depletion illuminates the temporal dynamics of the NMD-regulated human transcriptome” 🧬🧫
15.09.2025 16:12 — 👍 4 🔁 1 💬 0 📌 0

Carien Niessen elected new Vice-Rector for Research at the University of Cologne
The University Election Assembly elected Professor Dr Carien Niessen for the office of Vice-Rector for Research. The cell biologist is particularly committed to promoting excellent research at the uni...
🎉 Congratulations to our new Vice-Rector for Research, Professor Dr. Carien Niessen! 🙌
The University Council has elected Professor Niessen as Vice-Rector for Research to support the advancement of excellent research at the #UniCologne. 🎓
Read more 👇
uni.koeln/9FCEC
@cecad.bsky.social
10.09.2025 10:34 — 👍 11 🔁 3 💬 0 📌 0
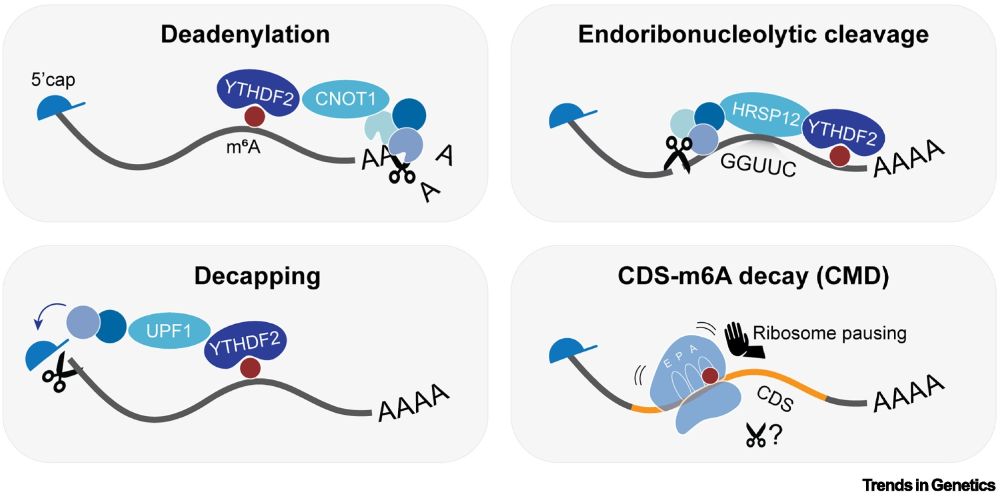
Figure 2. Overview of N6-methyladenosine (m6A) decay pathways. m6A acts as a destabilizing mark on mRNA.
"m6A in the coding sequence: linking deposition, translation and decay"
by Kathi Zarnack (@zarnack-group.bsky.social) & colleagues
"This review explores [m6A in] mRNA decay, with a focus on the newly discovered CDS–m6A decay (CMD) pathway..."
Read it here:
authors.elsevier.com/sd/article/S...
08.07.2025 12:07 — 👍 30 🔁 16 💬 0 📌 1
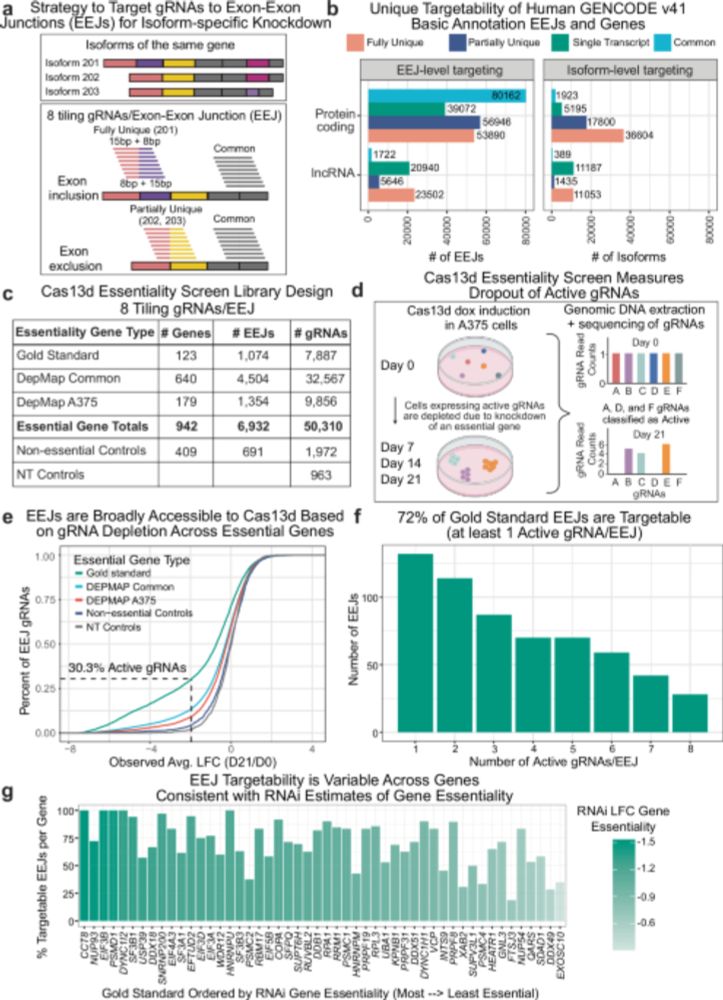
GENCODE - Home page
The latest human GENCODE releases for Human (49) and Mouse (M38), have just been released. They're available via gencodegenes.org and visible in @ensembl genome browser as part of Ensembl 115 ensembl.org
03.09.2025 11:16 — 👍 8 🔁 4 💬 1 📌 0

A conserved viral RNA fold enables nuclease resistance across kingdoms of life
Abstract. Viral exoribonuclease-resistant RNA (xrRNA) structures block cellular nucleases to produce subgenomic viral RNAs during infection. High sequence
Our study from the Steckelberg lab is out now in @narjournal.bsky.social !! We dove into a new nuclease-blocking viral #RNA structure important for viral infection, which provided evidence of similar structure-based strategies across diverse viral families. academic.oup.com/nar/article/...
02.09.2025 18:58 — 👍 22 🔁 10 💬 2 📌 2
🚨🚨🚨
Please RT!
We're looking for a postdoc to join an exciting joint project between our lab @upf.edu & @crg.eu (Barcelona) and the Sander lab @mdc-berlin.bsky.social (Berlin) investigating how alternative splicing and microexons influences the maturation of pancreatic islets.
Deadline: 30/09/25👇
23.07.2025 14:27 — 👍 48 🔁 65 💬 1 📌 3
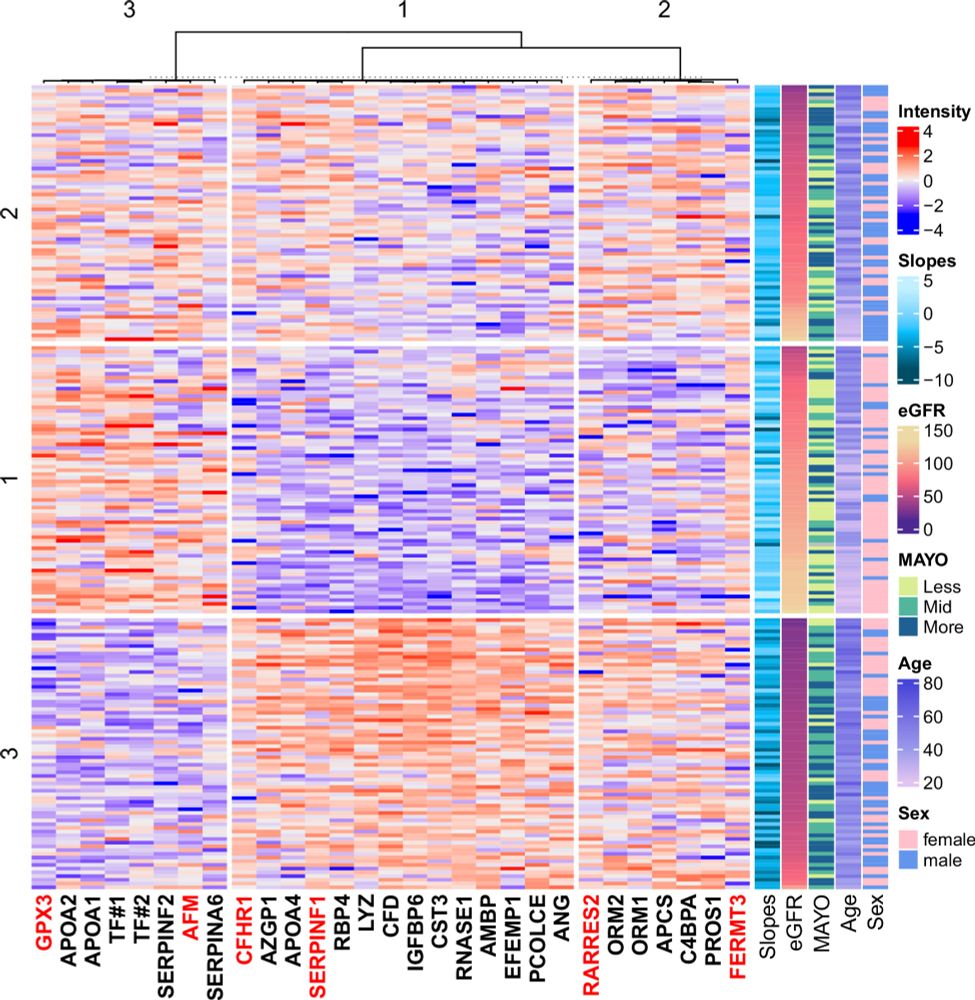
Developing serum proteomics based prediction models of disease progression in ADPKD
Nature Communications - Autosomal dominant polycystic kidney disease (ADPKD) is characterized by progressive cyst formation and loss of kidney function, yet prognostic biomarkers remain limited....
1/ 🧬 Exciting news! Our group just published a paper in Nature Communications on using serum proteomics to predict disease progression in #ADPKD (Autosomal Dominant Polycystic Kidney Disease) 🔬 — enhancing risk stratification beyond currently available models 🎯 rdcu.be/exbNq
21.07.2025 16:12 — 👍 8 🔁 3 💬 2 📌 0

Home
Hi #RNAsky, #ChromatinSky!
It is a pleasure to announce that the @ox.ac.uk RNA & transcription club is now on Bluesky. Follow us to stay in touch with everything RNA and transcription from gene regulation to disease. Join us here: rna.web.ox.ac.uk/home
18.07.2025 09:46 — 👍 11 🔁 5 💬 1 📌 0

Congratulations to our biochemistry professor Utz Fischer: he has been elected to @embo.org ! His research focuses on the macromolecular machines of the cell. 📷 Christoph Weiss www.uni-wuerzburg.de/en/news-and-...
01.07.2025 12:03 — 👍 12 🔁 2 💬 0 📌 1
Our latest study is now out in @narjournal.bsky.social: we identify a post-transcriptional bottleneck in interferon gamma signaling that hinges on proper maturation of JAK2 mRNA.
Paper: academic.oup.com/nar/article/...
01.07.2025 08:22 — 👍 8 🔁 4 💬 1 📌 1

🆕 publication! Cells don’t just fight infection - they fine-tune their immune response. A new study from the Versteeg lab in @narjournal.bsky.social led by Adrian Söderholm uncovers how RNA processing of JAK2 helps regulate immune signals ➡️ tinyurl.com/2py27c8u
@univie.ac.at
@meduniwien.ac.at
01.07.2025 06:28 — 👍 16 🔁 5 💬 3 📌 2
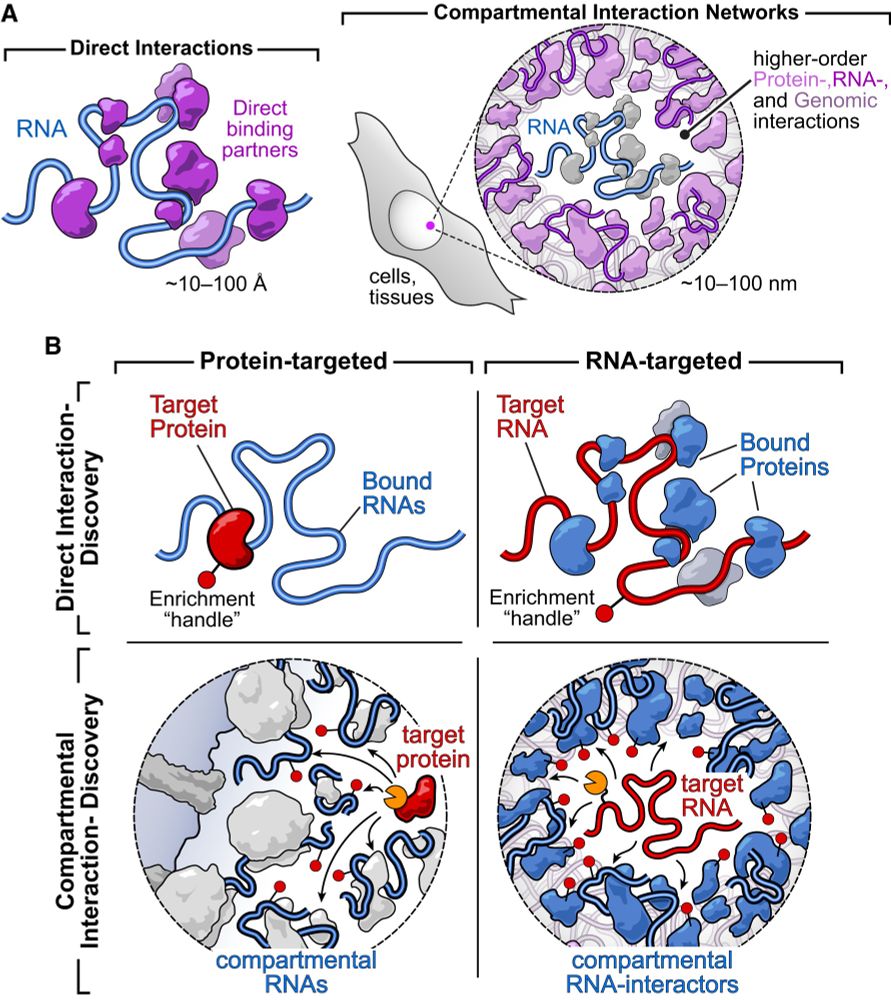
Hello! Ever wonder what's "talking to" your favorite transcript, but were too scared to ask? In our review in @cp-cellreports.bsky.social, @mardakheh.bsky.social and I highlight new RNA-focused tools for discovering RNA interactions across organizational scales. Checkit!
tinyurl.com/ydn6e3ac
30.06.2025 21:15 — 👍 116 🔁 48 💬 3 📌 5

Lexogen Goodie Bag
Feeling grateful to have received a fun goodie bag from #Lexogen for participating in their #RNA salon survey - special shoutout for the super cool sport shirt! 🎁🧬 Big thanks to the @rhinelandrna.bsky.social as well—always a pleasure to be part of this vibrant RNA community! #RNAsky
27.06.2025 09:58 — 👍 3 🔁 1 💬 1 📌 0
Contributing to a better world through research into the fundamental processes of living cells, tissues and organisms. The Hubrecht Institute is a research institute of KNAW and is affiliated with the UMC Utrecht.
Wandering around the complexities of genome regulation and anything connected to it 🧬
A dedicated fan of smiling out loud!
Currently at Sorbonne Université
RNA biologist 🧬 | Postdoc @zarnack-group in JMU Würzburg | From wet lab roots to computational biology
Abteilung für Molekulare und Medizinische Virologie
der Ruhr-Univeristät Bochum
https://virologie-bochum.de
RNA technologies. hesselberthlab.org
Husband. Father. Postdoc @ Svejstrup lab in Copenhagen studying transcription and DNA damage | EMBO & MSCA fellow
Jarocho de corazón, biochemist by training 🇲🇽
Svetlana Gorokhova, M.D., Ph.D., is working in Marseille Medical Genetics (@univamu) and @aphm-chu-marseille in Marseille, France, remote member of NNDCS @NIH @NINDSnews
Columbia PhD candidate studying viral RNA structure and degradation
Exploring RNA & RBPs in cancer: from cell edges to the chromatin. Microscopy lover, proteomics newbie. Likes computer sciences, physics & photography. Also an illustrator and graphic designer for Science and Biology (see: @STEMDORADOscimag)
I enjoy playing with family, reading science fiction and history, gardening, board games, and jazzercise. I'm a father, husband, Navy veteran, and graduate student studying computer science at George Mason University.
Mastodon: https://scicomm.xyz/@David
Postdoc at MD Anderson: molecular epi + statistical genomics of human development & cancer | PhD, Penn State: functional genomics in social insects.
Opinions are my own.
https://seantbresnahan.com
https://bhattacharya-lab.com
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
PhD in Biology 🔬 Amateur Runner 🏃♀️ Always learning something new 🌍 📖
The Oxford RNA Club is a dynamic community of researchers focused on all aspects of RNA biology, from gene regulation to disease. We host in-person monthly seminars, and larger events throughout the year. Join us here: https://rna.web.ox.ac.uk/home
Excellent science for tomorrow's plants.
Imprint: https://www.ceplas.eu/en/contact/imprint
#UniKöln #UniCologne // Impressum: http://ukoeln.de/3JGDD
Mastodon ➡ @UniKoeln@Wisskomm.social
Computational Biology group located at Universtiy of Würzburg, Germany. We work on #RNA_splicing #RNA_modification #RNA_transport #RNA_stabiliy #RBPs_in_Disease #DNA_replication #DNA_double_strand_break
machine learning and functional/statistical genetics. Associate Prof @Columbia and Core Faculty @nygenome. he/him/his. https://daklab.github.io/