
I am delighted to finally share our new preprint exploring the role of RBMX and its retrocopies in neurodevelopment:
👉🏻Read the full preprint here:
www.medrxiv.org/content/10.1...
Below are the key findings 👇🏻
@sgorokhova.bsky.social
Svetlana Gorokhova, M.D., Ph.D., is working in Marseille Medical Genetics (@univamu) and @aphm-chu-marseille in Marseille, France, remote member of NNDCS @NIH @NINDSnews

I am delighted to finally share our new preprint exploring the role of RBMX and its retrocopies in neurodevelopment:
👉🏻Read the full preprint here:
www.medrxiv.org/content/10.1...
Below are the key findings 👇🏻
Great start of the World Muscle Society #WMS2025 meeting in Vienna - an impressive wheelchair soccer match during the opening ceremony and Society’s 30y anniversary cake this morning! Now to the science with the first session on multisystemic alterations in muscle disease www.wms2025.com
08.10.2025 08:03 — 👍 0 🔁 0 💬 0 📌 0
🧐 This is how quality control works in our cells 🧬
Researchers from the #unicologne and the @mdc-berlin.bsky.social have cracked the code of a crucial mechanism of cellular quality control in humans. 👇
uni.koeln/RRCLZ

After our study on RNU4-2 and RNU5B-1 published in May (Nava et al, Nature Genetics 2025), I am excited to share our new preprint reporting dominant and recessive variants in RNU2-2 as a frequent cause of developmental and epileptic encephalopathy (DEE).
📄 www.medrxiv.org/content/10.1...
There is a new term that describes a translated region - "translon". This replaces multiple terms, such as ORF, CDS, non-canonical ORF etc.
07.09.2025 20:32 — 👍 0 🔁 0 💬 0 📌 0There's a new nomenclature for a translated region - "translon". This replaces multiple other terms, such as ORF, CDS, non-canonical ORF etc.
07.09.2025 20:26 — 👍 0 🔁 0 💬 0 📌 0

DMD duplications on carrier screening are not always pathogenic. Many are interspersed and benign. Assuming tandem equals disease risks misclassification. Long read sequencing reveals structure and can prevent unnecessary interventions. bit.ly/3HI4b2v
28.08.2025 00:02 — 👍 0 🔁 2 💬 0 📌 0This is a great reminder of the pitfalls of precomputed SpliceAI scores.. and a solution to quickly identify the variants that need to be re-annotated with SpliceAI github.com/Computationa...
29.08.2025 11:39 — 👍 1 🔁 0 💬 0 📌 0Also, check out the News & Views summarizing our findings, written by Youjia Guo & @themodzlab.bsky.social 🤩🤩
www.nature.com/articles/s41...
Excited to launch our AlphaGenome API goo.gle/3ZPUeFX along with the preprint goo.gle/45AkUyc describing and evaluating our latest DNA sequence model powering the API. Looking forward to seeing how scientists use it! @googledeepmind
25.06.2025 14:29 — 👍 219 🔁 82 💬 5 📌 10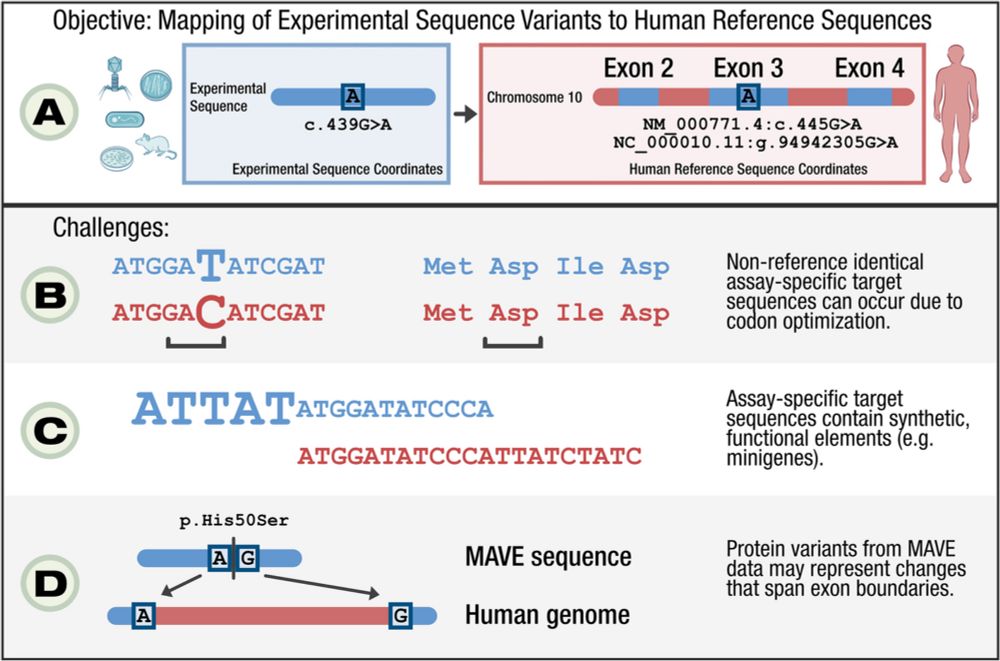
We are pleased to announce the publication of our manuscript "Mapping MAVE data for use in human genomics applications" in Genome Biology! 🧵 1/3 genomebiology.biomedcentral.com/articles/10....
25.06.2025 22:52 — 👍 6 🔁 5 💬 1 📌 0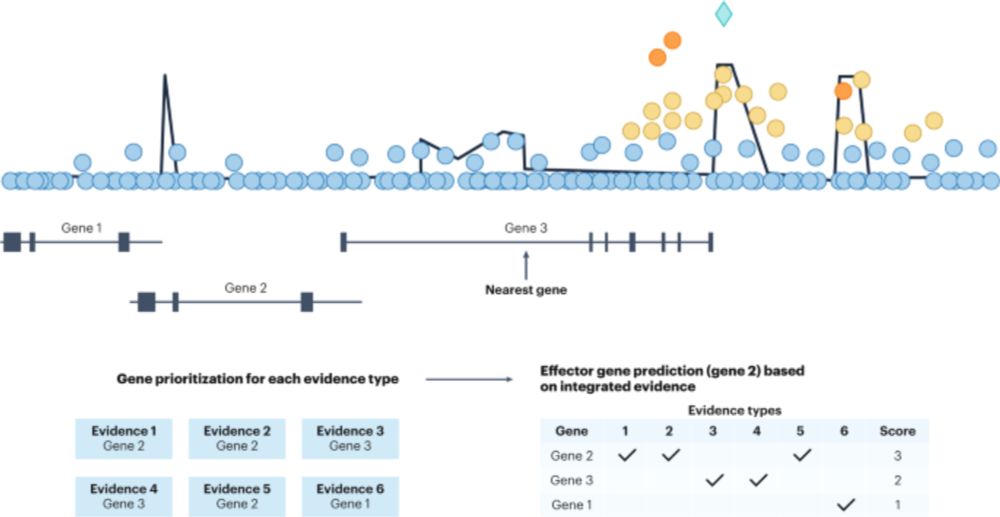
🚨Interested in which genes are the true effectors behind the scenes of the #gwas results? 🚨
This new publication in Nature Genetics, a collaboration between @broadinstitute.org @ebi.embl.org @opentargets.org focuses on approaches to predicted effector gene reporting tinyurl.com/98yxy4hf (1/3) 👇
Thanks to this new exome sequencing study, the number of genes with possible "human knockouts" goes up to 2,991!
13.06.2025 21:50 — 👍 0 🔁 0 💬 0 📌 0
We're super proud to see our study showing utility of proteomics in ultra-rapid variant prioritisation for suspected mito and other rare diseases out in Genome Medicine (rdcu.be/endwE). Too many amazing collabs to thank, so here are the big ones @daniellahock.bsky.social @thorburnmito.bsky.social!
23.05.2025 00:57 — 👍 28 🔁 11 💬 1 📌 5
Our recent paper is out: Distinct rates of VUS reclassification are observed when subclassifying VUS by evidence level buff.ly/Z0DcO9Q If you don't have access, our preprint is here buff.ly/z8UvVtY This paper emphasizes the critical benefit of VUS subclassification for physicians and patients.
13.04.2025 10:46 — 👍 17 🔁 5 💬 0 📌 0
I am delighted to share with you the news that our shiny new paper has hit the shelves in Genome Medicine!!
link.springer.com/article/10.1...
Key points (A 🧵):
These new SGE (saturation genome editing) scores will definitely be useful to resolve the RNU4-2 VUSes in our patients!
11.04.2025 10:52 — 👍 4 🔁 0 💬 0 📌 0🚨 Big news at #ACMG2025! 🚨
Today we’re announcing global democratization of deidentified allele count + frequency data with population breakdown from the first ~250k short-read WGS in All of Us designed to plug straight into clinical workflows. It is ~1.1 billion unique variants! 🧬💡
🧵 (1/4)
DECIPHER version 11.30 has been released. See the new features at www.deciphergenomics.org #variantinterpretation
21.03.2025 12:18 — 👍 7 🔁 5 💬 0 📌 1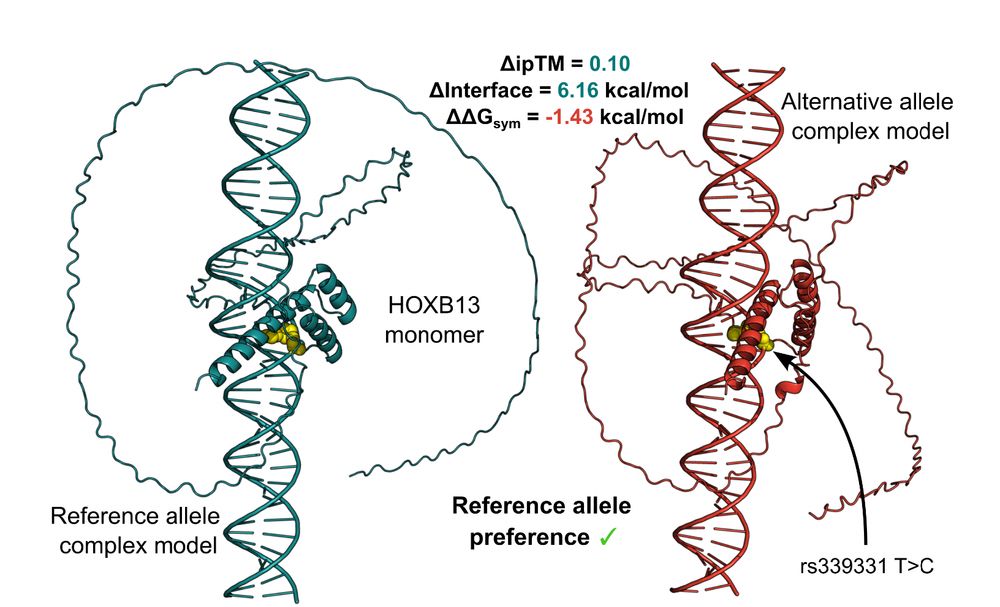
Unpicking non-coding genetic variation: Structure-guided modelling holds promise for evaluating how single nucleotide variants affect transcription factor binding. www.biorxiv.org/content/10.1.... @uoe-igc.bsky.social
21.03.2025 10:22 — 👍 16 🔁 8 💬 0 📌 0