I'm very excited to share something I've been working on off-and-on for a long time now: a new blog about genotype-phenotype landscapes! The first post is a Gödel-Escher-Bach-style dialogue to introduce the topic. If you like it please share/repost! open.substack.com/pub/topossib...
27.07.2025 20:18 — 👍 11 🔁 5 💬 0 📌 0

Grateful to be talking at #Evol2025! Will be presenting on how long Nanopore reads need to be in order to accurately call structural variants in Drosophila at the population level. Talk is at 3pm on Saturday in the Genomics III section. If you can’t make it get in touch!
20.06.2025 22:32 — 👍 13 🔁 3 💬 0 📌 0
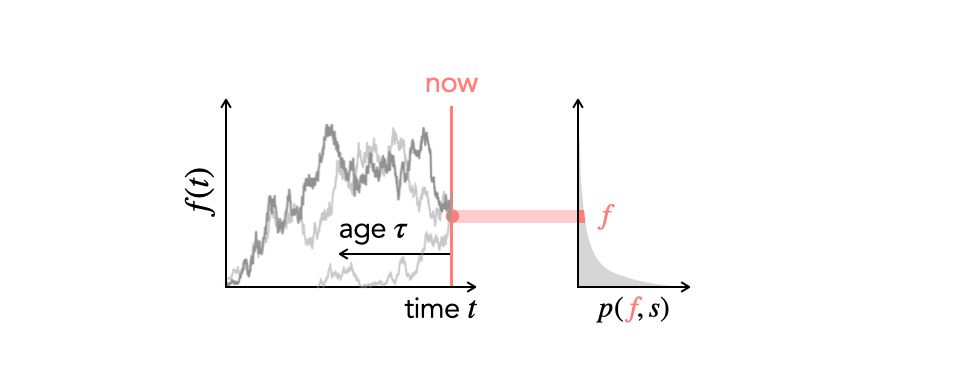
A schematic illustrating the allele frequency trajectories that contribute to a slice of the site frequency spectrum.
Looking forward to #evolution2025! I will be talking about how time-varying demography and selection shape the site frequency spectrum — Saturday at 4:15 pm, Population Genetics Theory IV. Come say hi if you are around!
20.06.2025 16:17 — 👍 21 🔁 4 💬 1 📌 1
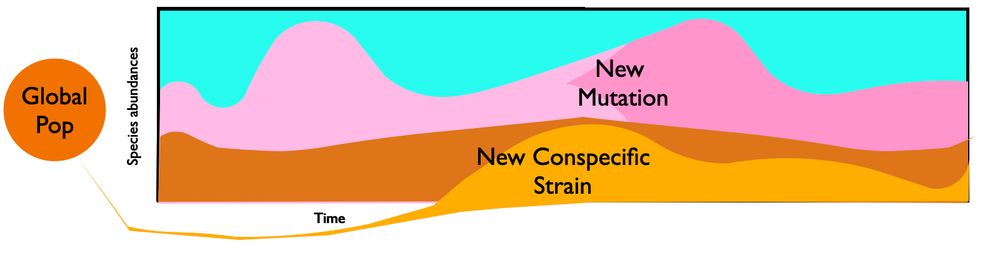
Mueller diagram of ecological and evolutionary dynamics in a gut microbiome. Species fluctuate in abundance and accumulate new mutations. New conspecific strains also colonize from the global population.
Super excited for #Evol2025! I will be sharing some cool work on selection in the gut microbial communities at 9:30 am on Monday in the Evolutionary Ecology I session (Parthenon 2)!! Looking forward to chatting with folks as well!
20.06.2025 16:35 — 👍 23 🔁 6 💬 1 📌 1
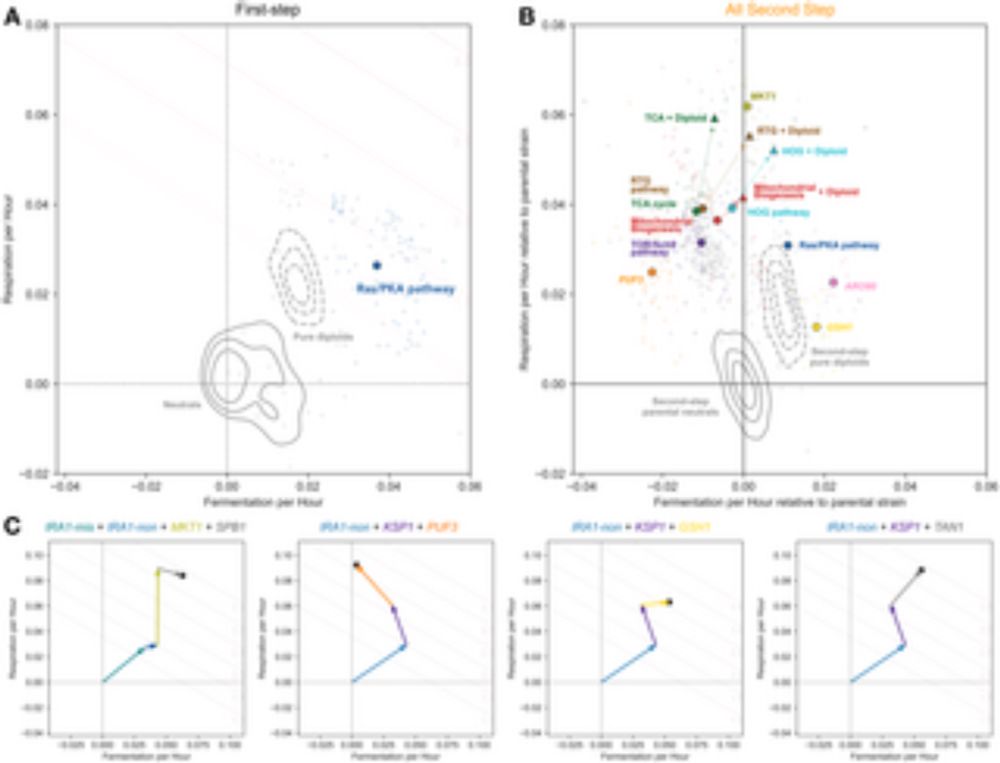
I think this emphasizes the need for more concrete models of pleiotropy to help us know when these verbal models should even apply in principle (let alone in expts like @oliviamghosh.bsky.social's). In that vein, was very excited about the new work that @djhelam1.bsky.social talked about at NITMB
20.06.2025 16:27 — 👍 3 🔁 1 💬 1 📌 0
Thank you for all your questions! I think they really point at some of the important challenges in interpreting this data.
20.06.2025 19:21 — 👍 2 🔁 0 💬 0 📌 0
I think it would be super cool to explore some of these assumptions and models theoretically, but (spoiler alert) we don't actually find evidence for the pleiotropic expansion model in our data! So perhaps these assumptions are not borne out in reality (in our system at least).
20.06.2025 13:34 — 👍 1 🔁 1 💬 0 📌 1
For the purposes of our paper, we are using it as a catch all for models in which the evolution condition is "special" from this top-down perspective. Specific mechanisms that would give rise to this are pretty subtle, as we have discussed here!
20.06.2025 13:34 — 👍 0 🔁 0 💬 1 📌 0
I think this raises the point that this pleiotropic expansion model is actually a little bit tricky to implement concretely, and definitely requires some additional assumptions.
20.06.2025 13:34 — 👍 0 🔁 0 💬 2 📌 0
this might mean that actually to make this model work, you do need some qualitative difference in E1 and E2 from the organism's perspective! ie how many traits are already optimized in each, and how those overlap with each other.
20.06.2025 13:34 — 👍 0 🔁 0 💬 1 📌 0
In E2, these yellow traits may not be optimized, so they are likely to be affected by adaptive mutants in E2. Then, when measured in E1, they will show up as additional detectable traits, as long as green traits were also not optimized in E2.
20.06.2025 13:34 — 👍 0 🔁 0 💬 1 📌 0
So you can imagine a scenario where there are certain traits that are already optimized in E1 (let's say these are the yellow traits). No adaptive mutant will affect these traits because changing them would decrease fitness (ignoring subtleties around the net fitness effect given multiple traits)
20.06.2025 13:34 — 👍 0 🔁 0 💬 1 📌 0
There are some implicit assumptions that must be true for this pleiotropic expansion model to work. First, just to clarify, the fact that a box is colorful does not mean it has a positive effect on fitness, it just means it is relevant. So a mutant's affect on a trait can be good in E1, bad in E2
20.06.2025 13:34 — 👍 0 🔁 1 💬 1 📌 0
You are bringing up an interesting (and somewhat subtle) point! I think if I understand your confusion properly you are saying that if there were these yellow traits that mattered to fitness in E1 all along, why didn't the mutants evolved in E1 ever affect them?
20.06.2025 13:34 — 👍 1 🔁 0 💬 2 📌 0
We expand on this more in the new text, so feel free to check it out! Also happy to chat more offline if it is still not clear.
19.06.2025 14:38 — 👍 0 🔁 0 💬 0 📌 0
So by that argument, if we looked at mutants evolved in E2, we would expect them to affect few traits in E2, and then many traits in E1.
19.06.2025 14:38 — 👍 0 🔁 0 💬 2 📌 0
But the "pleiotropic expansion" model says that the home environment is special. In the evolution environment, these mutants are *conditionally* low-dimensional.
19.06.2025 14:38 — 👍 0 🔁 0 💬 1 📌 0
On the assumption that mutations affect many traits and are generically pleiotropic, in most environments they will appear so, and hence we would discover a relatively "high" dimensional space by observing their fitness variation around that environment.
19.06.2025 14:38 — 👍 0 🔁 0 💬 1 📌 0
But the basic idea of what you said is right – there is no qualitative difference between environments 1 and 2. Instead, the "low-dimensionality" of home mutants is more of a statement about ascertainment bias.
19.06.2025 14:38 — 👍 0 🔁 0 💬 1 📌 0
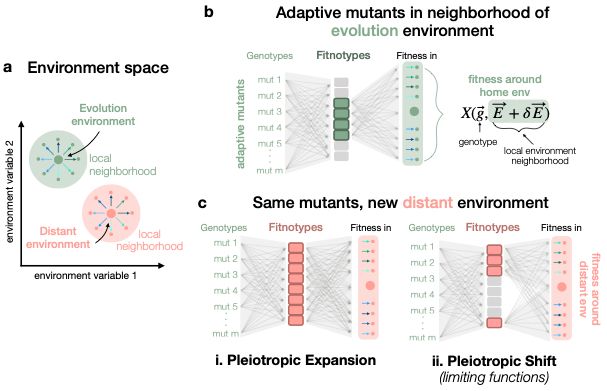
Thanks for taking a look! We actually have an updated v2 manuscript that I think clarifies our two competing hypotheses in figure 1:
www.biorxiv.org/content/10.1...
19.06.2025 14:38 — 👍 5 🔁 1 💬 1 📌 0

Excited to share SpaceBar - our new method for labeling and detecting clones with imaging-based spatial transcriptomics platforms! w/ Yael Heyman and @arjunraj.bsky.social www.biorxiv.org/content/10.1... 🧵
17.02.2025 18:44 — 👍 30 🔁 10 💬 2 📌 0
Professor, evolution of drug resistance, modeling, population genetics, coding, SF State University, mom, Dutch
Physics | Molecular Biology | Evolution.
Lecturer at UCL Physics.
Formerly:
Postdoc with @amurugan.bsky.social at the University of Chicago.
PhD with Madan Rao and Jitu Mayor at NCBS, Bangalore.
Evolutionary systems/cell biologist. EMBO and SNSF Postdoctoral Fellow with Dmitri Petrov and Dan Jarosz at Stanford. PhD with Andreas Wagner at the University of Zurich. Studying how molecular and cellular systems shape, and are shaped by, evolution.
Computational microbiologist. Senior scientist at @cemess.bsky.social, @univie.ac.at.
Microbial ecology, mostly of nitrogen cycle microbes, and data driven physiology.
Maintainer of the GlobDB genome database https://globdb.org
Systems Biology PhD Student @Harvard, Labs of Michael Desai @mmdesai.bsky.social & Michael Baym @baym.lol | Trinity College Cambridge Alum | Evolution & Microbes
Assistant Professor at UNC Chapel Hill
Assistant Professor @Biozentrum, University of Basel.
Jumbophage, Bacterial Immunity, Evolution
Evolutionary biologist; Lewis-Sigler Scholar @ Princeton. Formerly @ Harvard.
Postdoctoral researcher in Mia Levine’s Lab, University of Pennsylvania
Love all things Drosophila, evolution, running and Eastenders
Passionate about microbial symbiosis. Postdoc on🐻❄️longitudinal microbiome/virome at 🇨🇦UBC Zoology with @kayla-king.bsky.social, PhD 🇬🇧Oxford Zoology, BS 🇨🇳NPU. #poetry #art #coding #hiking lovers. #FirstGen #OpenScience. She/Hers.
Ph.D. Candidate at Stanford University working with Dr. Dmitri Petrov.
Assistant Professor of Physics at WashU working in areas of biophysics and soft matter.
I am interested in modelling biological system complexity relevant to ecology and evolution. Post doc at UC San Diego.
Website: djhelam.github.io
Google scholar: https://scholar.google.com/citations?user=RRxVJ6YAAAAJ&hl=en
Studying therapy resistance through the lens of complex systems, ecology, and evolution.
Current Jake Scott postdoc @ Cleveland Clinic
Former Kevin Wood graduate student @ Michigan
Theoretical ecologist, armchair philosopher
athma25.github.io
Asst. Prof. and DDLS fellow
Dept. Zoology, Stockholm University and SciLifeLab
🇦🇷 -> 🇫🇮 -> 🇸🇪
Evo-devo and dynamical systems
https://lisandromilocco.github.io/
assistant prof at University of Oregon. interested in pop gen, stat gen, human complex traits. also ELSI, metascience, ethics education, etc...
she/they. 🌈
roshnipatel.github.io
postdoc at Viral Ecology and Omics group, @microverse.bsky.social @uni-jena.de. Phages, metagenomics, evolutionary modeling, microbial communities, kendo.
Ph.D. student Landry Lab
gene duplication, cell signaling, protein evolution, full time member of the APOYG cult