See also the associated paper by Marcelo Torres, Fangping Wan & Cesar de la Fuente-Nunez
@delafuentelab.bsky.social
www.nature.com/articles/s41...
LauraMA
@laurama-2023.bsky.social
@laurama-2023.bsky.social
See also the associated paper by Marcelo Torres, Fangping Wan & Cesar de la Fuente-Nunez
@delafuentelab.bsky.social
www.nature.com/articles/s41...
Bacteria with archaella, who would have thought that this existed. Happy to share that it does 🎉and we just published it today in @natmicrobiol.nature.com 🎉🥳. Congrats to everyone involved! 🦠
17.09.2025 17:50 — 👍 23 🔁 3 💬 0 📌 0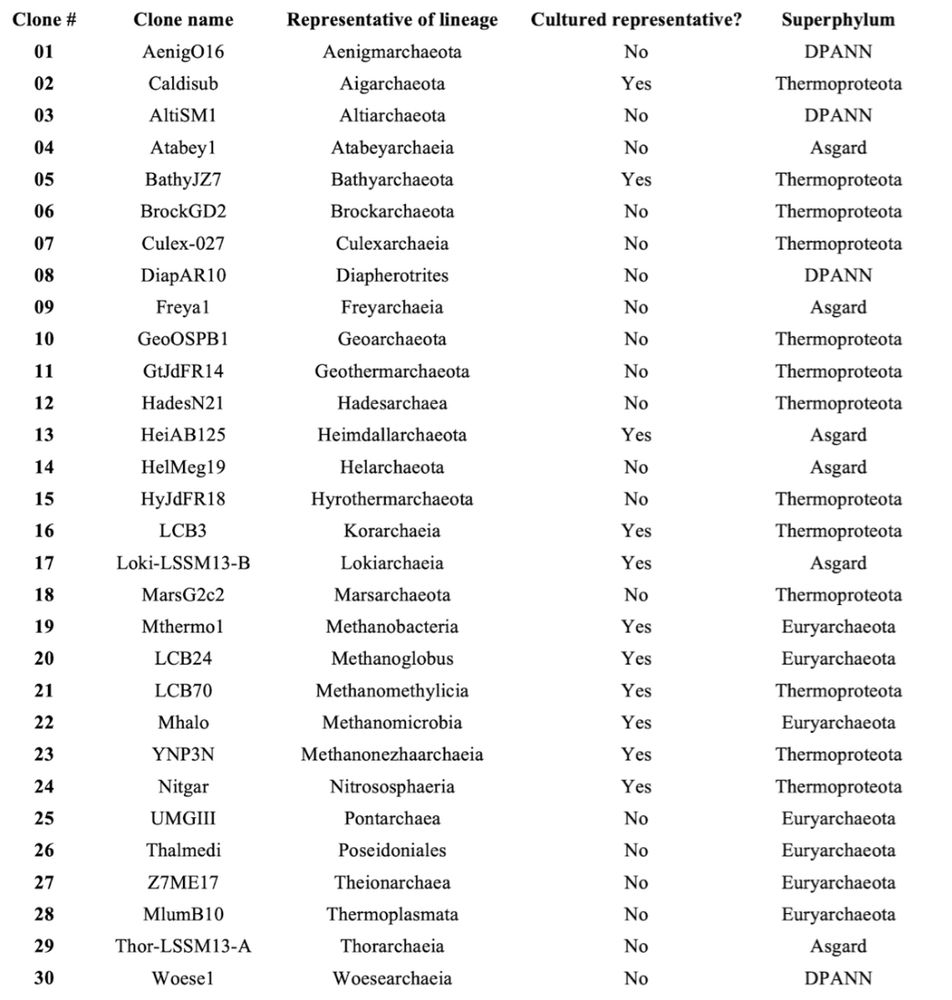
List of 30 CloneFISH cultures available for free (requester is asked to pay shipping fees).
Manuscript/resource alert #microsky 🦠 My lab offers the community a collection of 30 E. coli (CloneFISH) cultures, each carrying a plasmid for the heterologous expression of a (near) full-length 16S rRNA gene from one of 30 lineages of archaea, including 19 yet uncultured ones.
24.06.2025 13:53 — 👍 40 🔁 24 💬 5 📌 0
Joining the race to the bottom has never been cooler...
www.biorxiv.org/node/4580343...
www.biorxiv.org/content/10.1...
The Archaeacast! Yes, a podcast about #archaea!
by Priyanka Chatterjee, @cjhines.bsky.social, Alex Phillips, Theopi Rados & guests!
Sixth episode: Archaea and Us: Heroes or Villains?
rss.com/podcasts/arc...
@archaeapowerhour.bsky.social @archaeabio.bsky.social
#microsky

1/ Excited to share our latest work on gene transfer agents (GTAs) in Caulobacter crescentus, led by Emma Banks in collaboration with Pavol Bardy and Mai Nguyen from York!!! See a brief thread below.
shorturl.at/o6S1w
Thank you for reading and thanks to all co-authors!
18.04.2025 14:50 — 👍 0 🔁 0 💬 0 📌 0Therefore, in complex systems, proteins can serve multiple roles. Our findings highlight the importance of studying viral functions in their full biological context, and not just in overexpression setups. (10/n)
18.04.2025 14:50 — 👍 1 🔁 0 💬 1 📌 0We also analyzed 24 other viral AcrIII-1 homologs in archaea. Most are not expressed early and lack strong promoters, making them unlikely to act as Acrs in vivo. (9/n)
18.04.2025 14:50 — 👍 0 🔁 0 💬 1 📌 0Our data show cA4 influences viral fitness even without CRISPR systems, suggesting non-defense roles for this molecule that deserve further attention. (8/n)
18.04.2025 14:50 — 👍 0 🔁 0 💬 1 📌 0In fact, ring nuclease activity isn’t required for this advantage. SIRV2 gp37 appears multifunctional, since it also interacts with host proteins and may modulate cA4-linked pathways in ways unrelated to defense. (7/n)
18.04.2025 14:50 — 👍 0 🔁 0 💬 1 📌 0So, what does gp37 actually do? It binds and inhibits a host protein methyltransferase, giving the virus a fitness advantage that’s independent of CRISPR immunity. (6/n)
18.04.2025 14:50 — 👍 0 🔁 0 💬 1 📌 0We confirmed this by engineering early gp29 expression using an inducible promoter. This restored Acr activity and allowed viral replication under CRISPR pressure. (5/n)
18.04.2025 14:50 — 👍 0 🔁 0 💬 1 📌 0The reason for this is timing: gp37 is expressed mid/late in infection—too late to stop CRISPR-Cas, which acts early. Early expression is crucial for anti-CRISPR function. (4/n)
18.04.2025 14:50 — 👍 0 🔁 0 💬 1 📌 0We tested the native homolog, gp37 from SIRV2, in its natural host. But when gp37 is expressed from the virus genome, it shows no anti-CRISPR activity. (3/n)
18.04.2025 14:50 — 👍 0 🔁 0 💬 1 📌 0AcrIII-1 is a ring nuclease that degrades cA4, a key signaling molecule in type III CRISPR-Cas immunity.
Earlier studies showed that AcrIII-1 (like SIRV1 gp29) inhibits CRISPR, but only when overexpressed from plasmids in non-native systems. (2/n)

Our new paper on archaeal anti-CRISPRs is out in
@Nature (@SpringerNature)! 🔗 rdcu.be/eh1Ma
Using a native virus–host model (SIRV2–S. islandicus), we show that overexpression in foreign hosts can lead to false positives.
🧵(1/n) #CRISPR #archaea #antiCRISPR #virology

♀️🇲🇽🧪🦠 This paper about the challenges of being a female microbiologist in México is authored by four women central to my career:
my PhD supervisor Valeria Souza
my PhD committee member Gabriela Olmedo
my PhD colleagues Esmeralda Lopez and Ana Escalante
doi.org/10.1016/j.ti...
New paper! We analyzed gut #microbiomes of 250+ individuals (ages 19–109).
#Centenarians often show a high-methanogen “human cow” phenotype 🐄, with #Methanobrevibacter smithii dominance and youth-like archaeal profiles. #Archaea may stabilize networks in aging.
Link: rdcu.be/egk03
@archaeasky
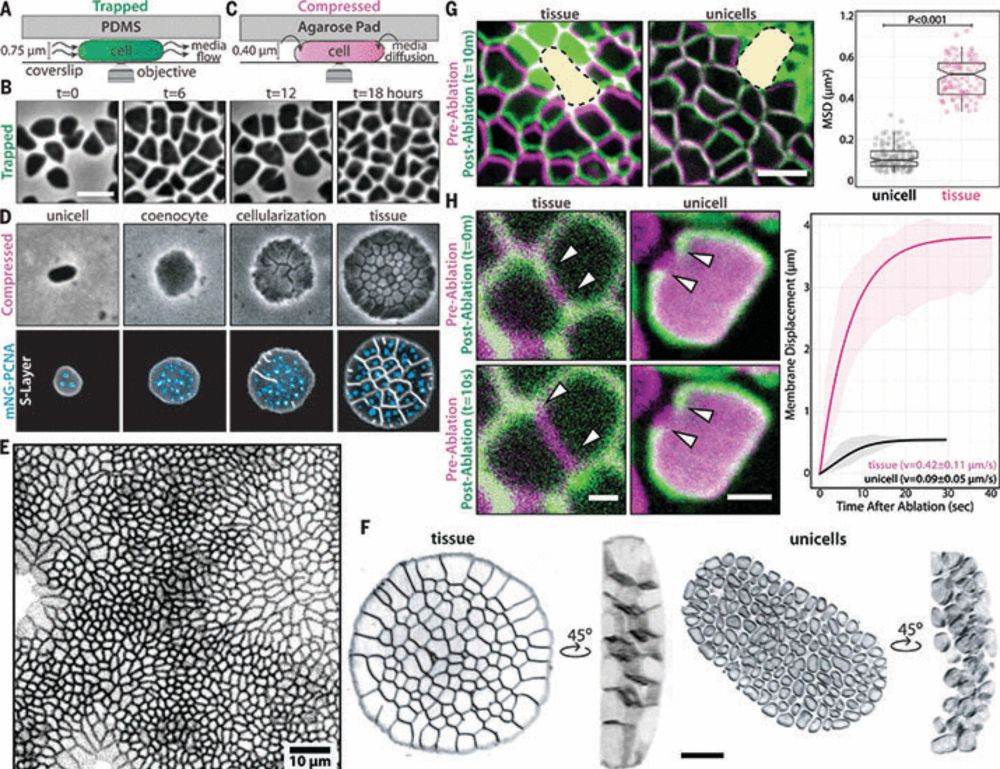
#weekendreading🔬📑
Tissue-like multicellular development triggered by mechanical compression in archaea
@science.org #microsky
www.science.org/doi/10.1126/...

From the @commsbio.bsky.social journal | Advancing archaeal research through FAIR resource and data sharing, and inclusive community building | #Bioinformatics #FAIR #OpenScience
#OpenAccess #OpenData 🧬 🖥️ 🧪 🔓
⬇️
www.nature.com/articles/s42...

Open PhD position! Do you want to work with the human #archaeome and understand the potential effect on the #immune system? Please check the call, including another exciting opening of @cdiener.com on #microbiome research www.medunigraz.at/doktoratsstu... #microbiomesky #archaeasky #microsky
13.02.2025 09:08 — 👍 24 🔁 12 💬 1 📌 2New preprint from our lab!
Story started long ago that took many years in the making.
Great collaboration with @archaellum.bsky.social & @tunglejic.bsky.social groups.
Generously funded by BBSRC @ukri.org & @leverhulme.bsky.social
How do #archaea segregate their chromosome?
#microsky #archaeasky
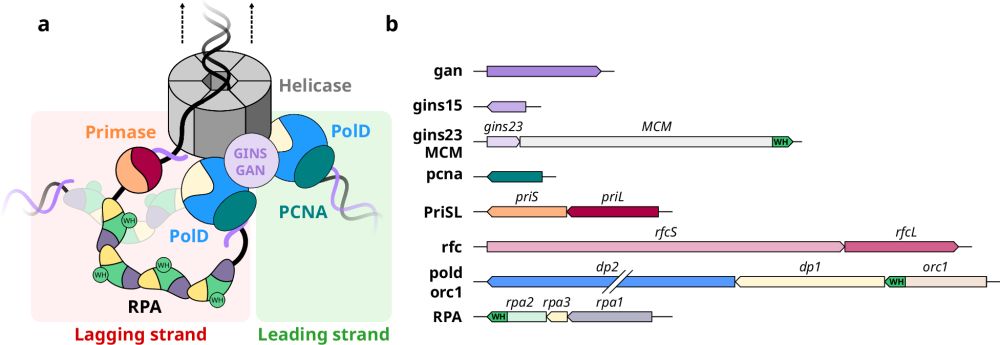
A new year, a new social network and a new article in @naturecomms.bsky.social ! It unveils how archaeal RPA orchestrates the recruitment of key players in 🧬 replication primase and polD through its WH domain, with structures of both complexes!!
rdcu.be/d6GAY