
Beeswarm plot of the prediction error across different methods of double perturbations showing that all methods (scGPT, scFoundation, UCE, scBERT, Geneformer, GEARS, and CPA) perform worse than the additive baseline.

Line plot of the true positive rate against the false discovery proportion showing that none of the methods is better at finding non additive interactions than simply predicting no change.
Our paper benchmarking foundation models for perturbation effect prediction is finally published 🎉🥳🎉
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
04.08.2025 13:52 — 👍 125 🔁 57 💬 2 📌 6
Thanks to all the coauthors, Eszter Varga, Daniel Dimitrov,
@juliosaezrod.bsky.social, László Hunyady and especially to first author Szilvia Barsi who led this project from start to finish! 7/7
15.07.2025 22:25 — 👍 0 🔁 0 💬 0 📌 0

Using an IO dataset of #nivolumab-treated renal cancer patients, we found that:
- #PD1 or #PDL1 expression didn't predict survival
- PD1 activity (from RIDDEN) did associate with survival.
Showing that receptor activity could be a more effective biomarker than expression. 5/n
15.07.2025 22:25 — 👍 0 🔁 0 💬 1 📌 0

RIDDEN’s receptor-specific signatures align with regulons of transcription factors downstream of receptors. 4/n
15.07.2025 22:25 — 👍 0 🔁 0 💬 1 📌 0

We benchmarked RIDDEN on various receptor perturbation datasets, including the recent in vivo Immune Dictionary data www.nature.com/articles/s41..., and found good predictive performance. 3/n
15.07.2025 22:25 — 👍 0 🔁 0 💬 1 📌 0

We used prior knowledge from omnipathdb.org & perturbation data from clue.io to build models for receptor activity inference. Instead of focusing on receptor expression, we predicted activity from the expression of receptor-regulated genes. 2/n
15.07.2025 22:25 — 👍 0 🔁 0 💬 1 📌 0

Our tool, RIDDEN (Receptor actIvity Data Driven inferENce) for predicting #receptor activity from #transcriptomics data is published in
PLOS Computational Biology journals.plos.org/ploscompbiol... 1/n
15.07.2025 22:25 — 👍 3 🔁 1 💬 1 📌 0
Thanks all the co-authors, Gema Sanz, @kristofszalay.com and especially Gerold Csendes, who led this project! 5/5
25.04.2025 21:49 — 👍 0 🔁 0 💬 0 📌 0

Several popular Perturb-seq based benchmark datasets lack heterogeneity, making it difficult to distinguish between strong and weak models. 4/5
25.04.2025 21:49 — 👍 0 🔁 0 💬 1 📌 0
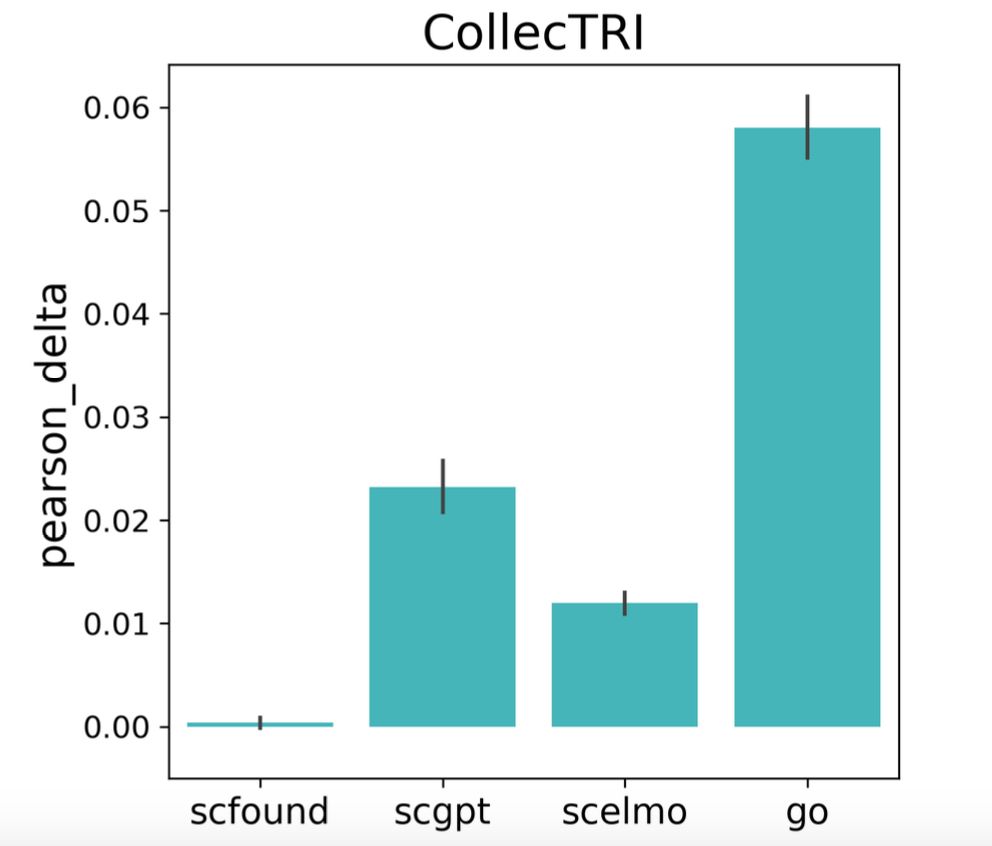
Gene embeddings from foundation models align more closely with gene regulatory networks than with signaling networks, which may underlie their weaker performance in perturbation tasks. 3/5
25.04.2025 21:49 — 👍 0 🔁 0 💬 1 📌 0

Even the most trivial baseline (mean of train samples) outperformed recent foundation models, while basic ML models using biologically meaningful features won by a large margin. 2/5
25.04.2025 21:49 — 👍 0 🔁 0 💬 1 📌 0

Single-cell foundation models, trained on large-scale scRNA-seq datasets, are increasingly used for post-perturbation RNA-seq prediction.
But how do they actually perform?
Our new paper from @turbine-ai.bsky.social is now out in BMC Genomics. bmcgenomics.biomedcentral.com/articles/10....
1/5
25.04.2025 21:49 — 👍 2 🔁 1 💬 1 📌 0

Hello #AACR25 !
25.04.2025 16:10 — 👍 5 🔁 0 💬 0 📌 0

PhD/Postdoc in Spatial Omics for Cardiology
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!
We have an opening for a PhD/Postdoc in Spatial Omics for Cardiology in Heidelberg 🫀 #job #academicjob #phd #postdoc #cadiology 👇
jobrxiv.org/job/heidelbe...
22.04.2025 10:28 — 👍 6 🔁 8 💬 0 📌 0

Pretraining virtual cells is useless
the way we do it now
Turns out the way we usually pre-train foundational cell models adds very little information to the system - definitely not enough to make drug effect predictions work. Not what I've expected.
#virtualcells #foundationalmodels #compbio
blog.turbine.ai/p/pretrainin...
16.04.2025 17:03 — 👍 2 🔁 2 💬 0 📌 0
easter holiday is coming: 2 revised versions submitted today
03.04.2025 22:48 — 👍 0 🔁 0 💬 0 📌 0
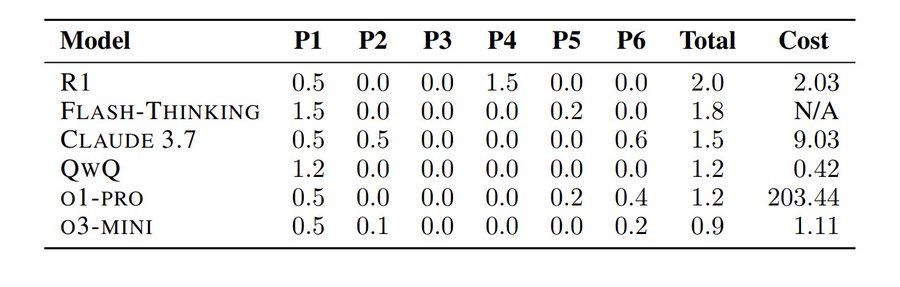
You’ve probably heard about how AI/LLMs can solve Math Olympiad problems ( deepmind.google/discover/blo... ).
So naturally, some people put it to the test — hours after the 2025 US Math Olympiad problems were released.
The result: They all sucked!
31.03.2025 20:33 — 👍 174 🔁 50 💬 9 📌 12
Does @iscb.bsky.social have a statement on this? We need to be there to support bioinformatics researchers.
30.03.2025 18:23 — 👍 23 🔁 9 💬 0 📌 0
based on some recent papers / presentations, the hill(s) I'll die on:
1) Raw IC50 values != good metrics for comparing cancer drug sensitivity
2) Raw TPMs / counts are meaningless for comparing gene expression similarity
3) Diverging color palettes need a meaningful midpoint.
30.03.2025 20:44 — 👍 1 🔁 0 💬 0 📌 0

Flier for the ICSB2025 conference in Dublin 5-9 October 2025
We will be hosting the International Conference on Systems Biology (ICSB) in Dublin this year – 5-9th October icsb2025.com/registration/ #ICSB2025
29.01.2025 16:50 — 👍 12 🔁 7 💬 0 📌 2
It's been over 24 hours since #AdventOfCode 2024 ended!
This AoC season:
- 273,313 users collected at least one star
- 3,654,949 stars were collected
Since 2015:
- 779 users have all 500 stars
- 1,077,226 users collected at least one star
- 23,170,305 were collected
26.12.2024 22:21 — 👍 270 🔁 20 💬 17 📌 0
holiday season is coming: 2 papers submitted today
10.12.2024 22:55 — 👍 1 🔁 1 💬 0 📌 1
Thanks to all the coauthors, Eszter Varga, Daniel Dimitrov, @juliosaezrod.bsky.social , László Hunyady and especially to first author Szilvia Barsi who led this project from start to finish! 6/6
09.12.2024 21:39 — 👍 0 🔁 0 💬 0 📌 0
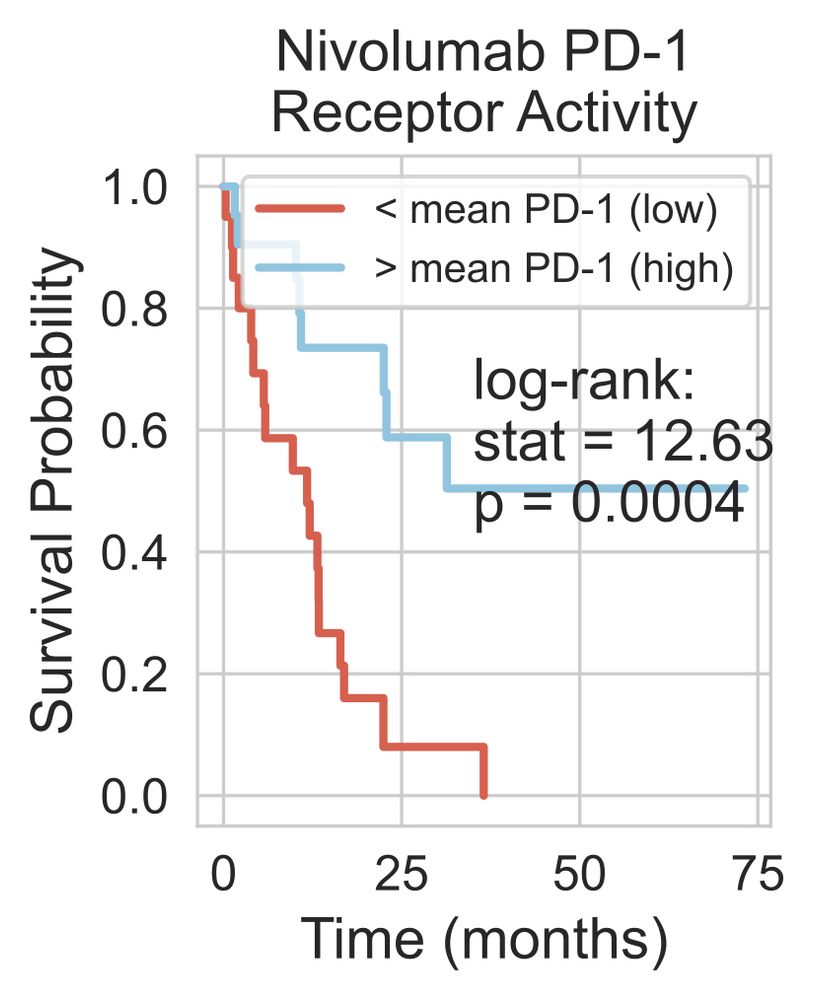
Using an IO dataset of nivolumab-treated renal cancer patients, we found that:
- PD1 or PDL1 expression didn't predict survival
- PD1 activity (from RIDDEN) did associate with survival.
This shows that receptor activity could be a more effective biomarker than expression. 5/n
09.12.2024 21:39 — 👍 0 🔁 0 💬 1 📌 0
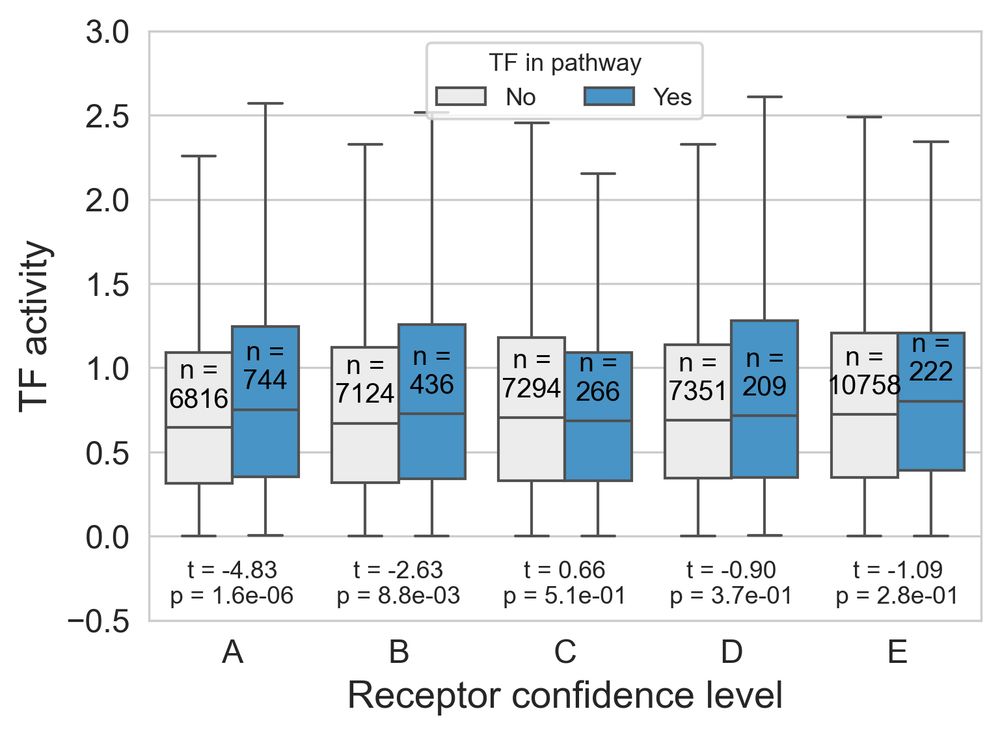
RIDDEN’s receptor-specific signatures align with regulons of transcription factors downstream of receptors. 4/n
09.12.2024 21:39 — 👍 0 🔁 0 💬 1 📌 0

We benchmarked RIDDEN on various receptor perturbation datasets, including the recent in vivo Immune Dictionary data nature.com/articles/s41..., and found good predictive performance. 3/n
09.12.2024 21:39 — 👍 0 🔁 0 💬 1 📌 0

We combined prior knowledge from OmniPath omnipathdb.org with LINCS L1000 perturbation transcriptomics clue.io to build models that infer receptor activity. Instead of focusing on receptor expression, we predict activity using the expression of receptor-regulated genes. 2/n
09.12.2024 21:39 — 👍 0 🔁 0 💬 1 📌 0
In Almouzni Lab at Institut Curie, a bioinformatician among H3 histone variants afficionados, exploring their role in shaping chromatin in pediatric brain cancers.
Mexican Historian & Philosopher of Biology • Postdoctoral Fellow at @theramseylab.bsky.social (@clpskuleuven.bsky.social) • Book Reviews Editor for @jgps.bsky.social • https://www.alejandrofabregastejeda.com • #PhilSci #HistSTM #philsky • Escribo y edito
Associate professor @ South China University of Technology. Interested in synthetic biology, genome editing, and directed evolution.
Web development, data management and visualizations. Consulting services for scientific and r&d communities. https://www.researchelements.org/
research professor
@semmelweishu, http://hegelab.org; exp: transmembrane proteins, transporters, 3D-bioinformatics, computational biology; father of 3
physicist, bioinformatics / biostats researcher at University of Heidelberg
Statistician and computational biologist; uw alum; jhu student. He/him. http://ekernf01.github.io
Mucosal Immunologist interested in the complexity of IBD. Professor @Karolinskainst, Wallenberg Fellow & editor @MucosalImmunol. Opinions are my own
Bioinformatician, Quadram & Earlham Institute, Norwich, loves when programs break .. not
Turbine is virtualizing experiments with AI to accelerate discovery and clinical decisions. By simulating how cells and tissues behave under treatment, Turbine helps pharma identify the right therapeutic ideas smarter and faster.
Data-driven & knowledge-based. Rooted in Hamburg, Germany.
Adipose and/or Systems Biology + Inflammation.
Account handled by π @LorenzAdlung.com
www.adlunglab.com
A career network featuring science jobs in academia and industry.
Visit our platform at www.science.hr
Plant scientist 🌱🦠🧫 🇲🇽 | Postdoc at the University of Zurich in the Cyril Zipfel and Pedro Beltrao groups, studying protein phosphorylation during plant–microbe interactions #EMBOfellow | @thesainsburylab.bsky.social alumni
MedTech software development, woodworking, history. Durham NC.
PMGC offers high-quality, cost-effective genomics services to academic & commercial clients worldwide. Specializing in DNA seq, epigenomics, panel-based, single-cell & spatial genomics. Get in touch for custom unique solutions for your research!
Researcher at Heidelberg University @saezlab.bsky.social | #ML and AI in #single-cell and #spatial #omics
Computational biologist | Omics data analysis | Post-doctoral researcher | University of Cologne
Defended PhD and current postdoc @MeauxJuliette.bsky.social lab @UniCologne. Plant physiological and genetic response to abiotic stresses |MicroRNAs are amazing. ECR @trr341.bsky.social |
https://www.linkedin.com/in/abdul363
We are a leading Cancer Research Institute within The University of Manchester, funded by Cancer Research UK (@cancerresearchuk.org)
https://www.cruk.manchester.ac.uk/
Elihu Professor of Biostatistics @yalesph.bsky.social--Cancer, Infectious disease, Evolutionary biology, Fungi, sometimes running





















